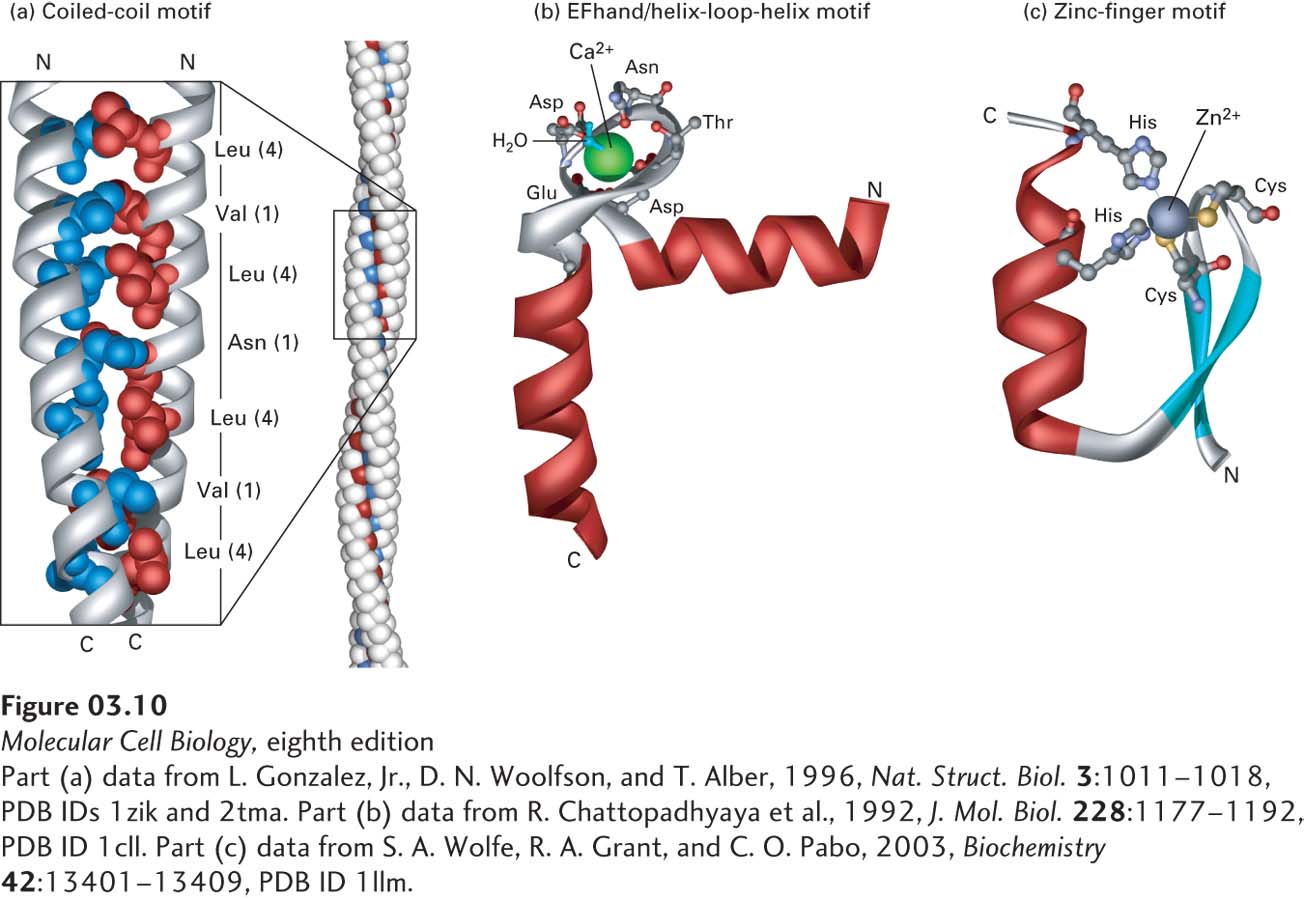
FIGURE 3- 10 Motifs of protein secondary structure. (a) This parallel two- stranded coiled- coil motif (left) is characterized by two α helices wound around each other. Helix packing is stabilized by interactions between hydrophobic side chains (red and blue) present at regular intervals along each strand and found along the seam of the intertwined helices. Each α helix exhibits a characteristic heptad repeat sequence with a hydrophobic residue often, but not always, at positions 1 and 4, as indicated. The coiled- coil nature of this structural motif is more apparent in long coiled coils containing many such motifs (right). (b) An EF hand, a type of helix- loop- helix motif, consists of two helices connected by a short loop in a specific conformation. This structural motif is common to many proteins, including many calcium- binding and DNA- binding regulatory proteins. In calcium- binding proteins such as calmodulin, oxygen atoms from five residues in the acidic glutamate- and aspartate- rich loop and one water molecule form ionic bonds with a Ca2+ ion. (c) The zinc- finger motif is present in many DNA- binding proteins that help regulate transcription. A Zn2+ ion is held between a pair of β strands (blue) and a single α helix (red) by a pair of cysteine residues and a pair of histidine residues. The two invariant cysteine residues are usually at positions 3 and 6, and the two invariant histidine residues are at positions 20 and 24 in this 25- residue motif.
[Part (a) data from L. Gonzalez, Jr., D. N. Woolfson, and T. Alber, 1996, Nat. Struct. Biol. 3:1011– 1018, PDB IDs 1zik and 2tma. Part (b) data from R. Chattopadhyaya et al., 1992, J. Mol. Biol. 228:1177– 1192, PDB ID 1cll. Part (c) data from S. A. Wolfe, R. A. Grant, and C. O. Pabo, 2003, Biochemistry 42:13401– 13409, PDB ID 1llm.]
[Leave] [Close]