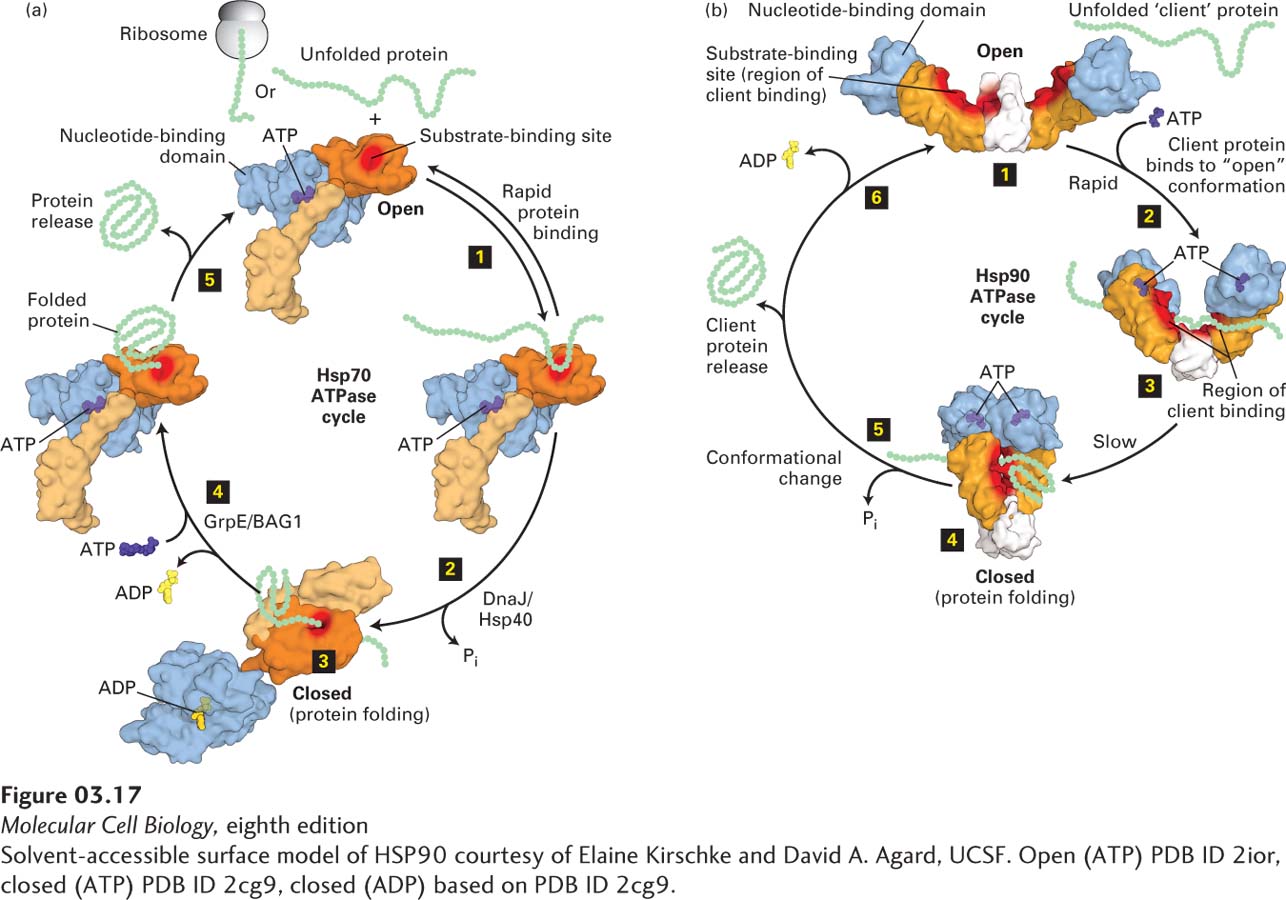
FIGURE 3- 17 Molecular chaperone– mediated protein folding. (a) Hsp70. Many proteins fold into their proper three- dimensional structures with the assistance of Hsp70 or one of several Hsp70- like proteins. These molecular chaperones transiently bind to a nascent polypeptide as it emerges from a ribosome or to a protein that has otherwise unfolded. In the Hsp70 cycle, an unfolded substrate protein binds in rapid equilibrium (step 1) to Hsp70’s substrate- binding site (red) in the open conformation of its substrate- binding domain (light and dark orange) when an ATP (purple) is bound at Hsp70’s nucleotide- binding domain (light blue). The substrate- binding domain comprises two subdomains (light and dark orange) that change relative positions and conformations during the cycle. Co- chaperone accessory proteins (DnaJ/Hsp40) stimulate the hydrolysis of ATP to ADP (yellow) that induces a large conformational change in the substrate- binding domain, resulting in the closed conformation, in which the substrate is locked into the substrate- binding domain; here proper folding is facilitated (steps 2 and 3). Exchange of ATP for the bound ADP, stimulated by other accessory co- chaperone proteins (GrpE/BAG1), converts the Hsp70 back to the open conformation (step 4), releasing the properly folded substrate (step 5) and regenerating the open conformation, which can then interact with additional substrates. (b) Three conformational states of the dimeric Hsp90 molecular chaperone thought to be involved in substrate (also called client) remodeling. Client proteins bind at the substrate- binding site (red surface) shared by the substrate- binding (orange) and C- terminal dimerization (white) domains and are thought to be remodeled in response to ATP binding and hydrolysis. The Hsp90 cycle begins when there is no nucleotide bound to the nucleotide- binding domains (light blue) and the dimer is in a very flexible, open configuration (step 1) that can bind a client. Rapid ATP binding leads to a conformational change (step 2) in which the nucleotide- binding domains and the substrate- binding domains move together (intermediate shown in step 3) into a closed conformation in which the nucleotide- binding domains are dimerized (step 4). The precise locations in Hsp90 at which clients bind apparently vary for different clients, but the binding surface, including the intersection of the substrate- binding domains and C- terminal dimerization domains (highlighted by red shading) binds a number of clients. ATP hydrolysis results in a conformational change in Hsp90 (step 5) that may include a highly compact form, folding of the client, and client protein release. The ADP- bound form of Hsp90 can adopt several conformations, including a highly compact form. Release of ADP (step 6) regenerates the initial flexible open state, which can then interact with additional clients. See E. D. Kirschke et al., 2014, Cell 157:1685 and M. Taipale, D. F. Jarosz, and S. Lindquist, 2010, Nat. Rev. Mol. Cell Biol. 11:515.
[Solvent- accessible surface model of HSP90 courtesy of Elaine Kirschke and David A. Agard, UCSF. Open (ATP) PDB ID 2ior, closed (ATP) PDB ID 2cg9, closed (ADP) based on PDB ID 2cg9.]
[Leave] [Close]