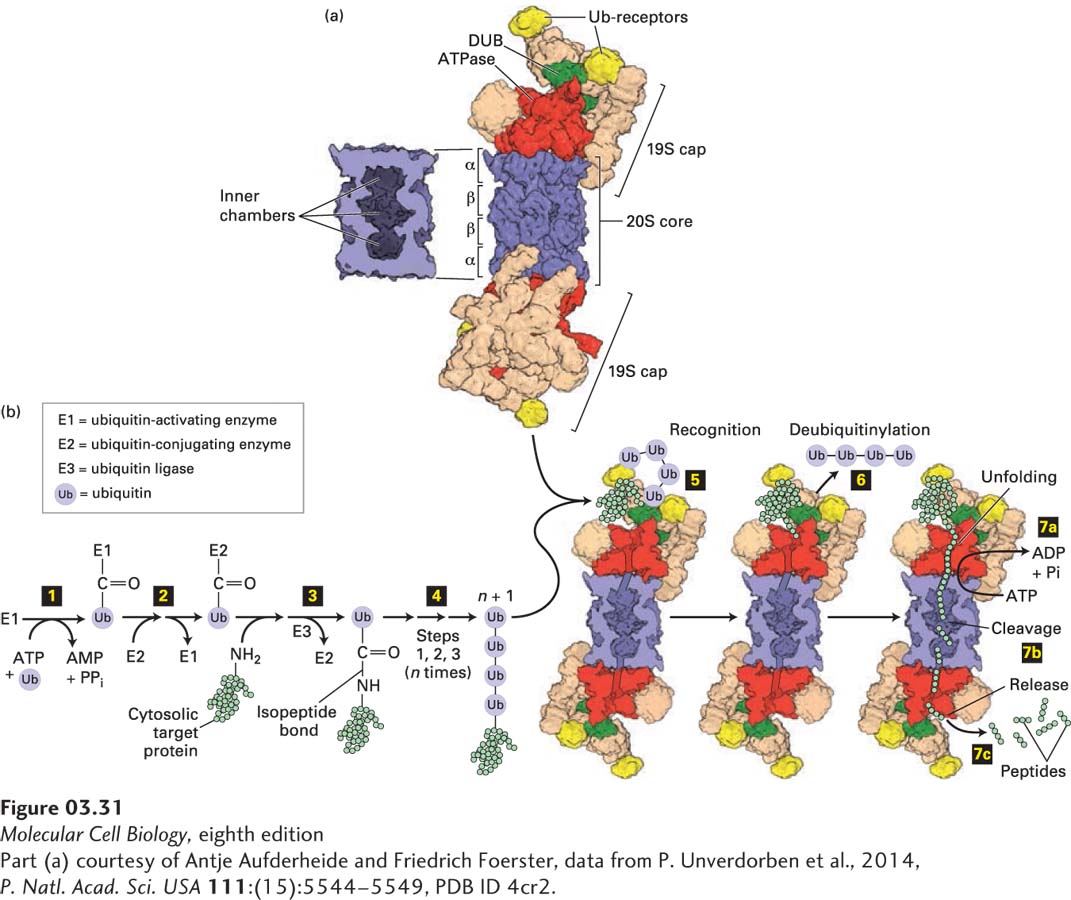
FIGURE 3- 31 Ubiquitin- and proteasome- mediated proteolysis. (a) Right: The 26S proteasome has a cylindrical structure with a 19S cap at one or both ends of the 20S core particle. The nineteen different subunits of the 19S cap (shown in multiple colors) include six AAA- ATPase subunits (Rpt1– 6, red), which assemble into a heterohexameric ring; two ubiquitin (Ub) receptors (Rpn10 and Rpn13, yellow); and a deubiquitinase enzyme (DUB, Rpn11, green), which forms a heterodimer with its evolutionarily related counterpart Rpn8. Moreover, the 19S cap contains scaffolding and other proteins (tan). The two 19S caps shown are facing in opposite directions relative to the plane of the page. The 20S core consists of four stacked heptameric rings (~110 Å diameter × 160 Å long), each containing either α (outer rings) or β (inner rings) subunits (blue). Left: Cutaway view of the 20S core, showing the inner chambers. Proteolysis occurs within the central inner chamber of the core formed by the β rings. (b) Proteins are targeted for proteasomal degradation by polyubiquitinylation. Enzyme E1 is activated by the ATP- dependent attachment of a ubiquitin (Ub) molecule (step 1) and then transfers this Ub molecule to a cysteine residue in E2 (step 2). Ubiquitin ligase (E3) transfers the bound Ub molecule on E2 to the side- chain –NH2 of a lysine residue in a target protein, forming an isopeptide bond (step 3). Additional Ub molecules are added to the Ub- modified target protein via isopeptide bonds to the previously added Ub by repeating steps 1 – 3, forming a polyubiquitin chain (step 4). The polyubiquitinylated target is recognized by Ub receptors in the proteasome’s 19S cap (step 5), and the Ub groups are removed by the deubiquitinase enzyme (step 6). In step 7 ATP hydrolysis enables the six protein (hexameric) ATPase subunits (red) to unfold the substrate and transfer the unfolded protein via a pore in the hexamer into the proteolysis chamber in the 20S core (step 7a), in some cases coordinately with step 6, and the protein is cleaved into short peptide digestion fragments (step 7b) that are then released (step 7c).
[Part (a) courtesy of Antje Aufderheide and Friedrich Foerster, data from P. Unverdorben et al., 2014, P. Natl. Acad. Sci. USA 111:(15):5544– 5549, PDB ID 4cr2.]
[Leave] [Close]