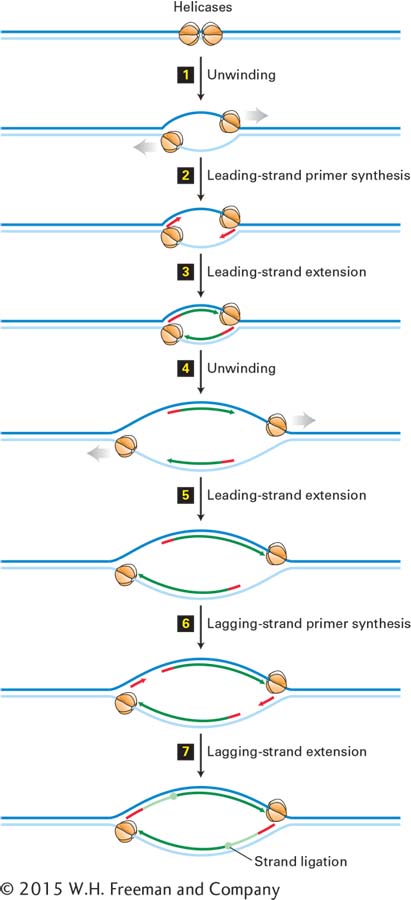
FIGURE 5- 32 Bidirectional mechanism of DNA replication. The left replication fork here is comparable to the replication fork diagrammed in Figure 5- 30 (although that figure also shows proteins other than large T- antigen, which are not shown here). Top: Two large T- antigen hexameric helicases first bind at the replication origin in opposite orientations. Step 1: Using energy provided by ATP hydrolysis, the helicases move in opposite directions, unwinding the parent DNA and generating single- stranded templates, which are bound by RPA proteins. Step 2: Primase– Pol α complexes synthesize short primers (red) base- paired to each of the separated parent strands. Step 3: PCNA- Rfc– Pol ε complexes replace the primase– Pol α complexes and extend the short primers, generating the leading strands (dark green) at each replication fork. Step 4: The helicases further unwind the parent strands, and RPA proteins bind to the newly exposed single- stranded regions. Step 5: PCNA- Rfc– Pol ε complexes extend the leading strands farther. Step 6: Primase– Pol α complexes synthesize primers for lagging- strand synthesis at each replication fork. Step 7: PCNA- Rfc– Pol δ complexes displace the primase– Pol α complexes and extend the lagging- strand Okazaki fragments (light green), which are eventually ligated to the 5′ ends of the leading strands. The position where ligation occurs is represented by a circle. Replication continues by further unwinding of the parent strands and synthesis of leading and lagging strands as in Steps 4–7. Although depicted as individual steps for clarity, unwinding and synthesis of leading and lagging strands occur concurrently.
[Leave] [Close]