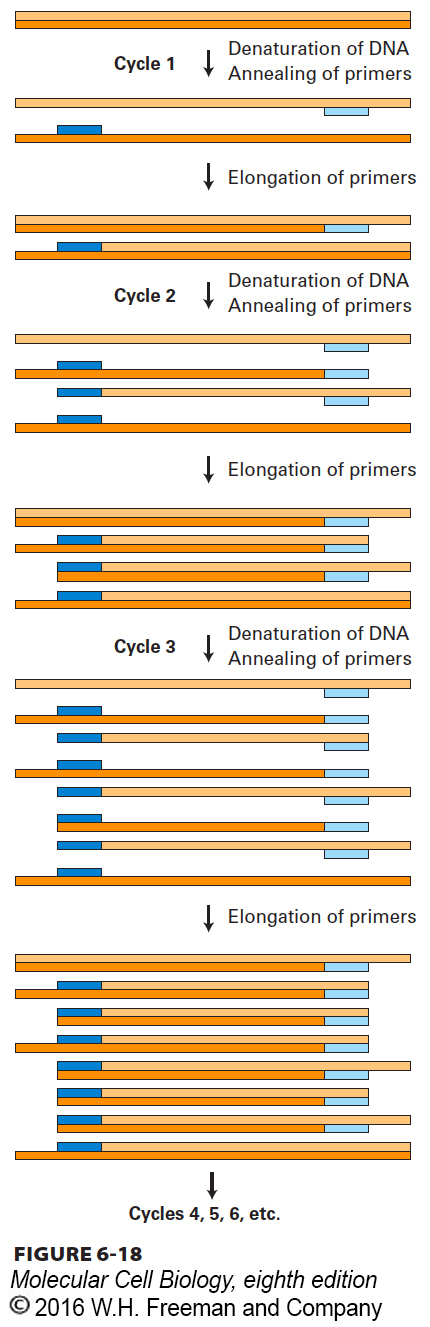
FIGURE 6- 18 The polymerase chain reaction (PCR) is widely used to amplify DNA regions with known flanking sequences. To amplify a specific region of DNA, an investigator chemically synthesizes two different oligonucleotide primers complementary to sequences of approximately 18 bases flanking the region of interest (shown here as light blue and dark blue bars). The complete reaction is composed of a complex mixture of double- stranded DNA (usually genomic DNA containing the target sequence of interest), a stoichiometric excess of both primers, the four dNTPs, and a heat- stable DNA polymerase known as Taq polymerase. During each PCR cycle, the reaction mixture is first heated to separate the strands and then cooled to allow the primers to bind to complementary sequences flanking the region to be amplified. Taq polymerase then extends each primer from its 3′ end, generating newly synthesized strands that extend in the 3′ direction to the 5′ end of the template strand. During the third cycle, two double- stranded DNA molecules are generated equal in length to the sequence of the region to be amplified. In each successive cycle, the target sequence, which anneals to the primers, is duplicated, so it eventually vastly outnumbers all other DNA sequences in the reaction mixture. Successive PCR cycles can be automated by cycling the reaction at timed intervals between a high temperature for DNA melting and a lower temperature for the annealing and elongation parts of the cycle. A reaction that cycles 20 times will amplify the specific target sequence 1- million- fold.
[Leave] [Close]