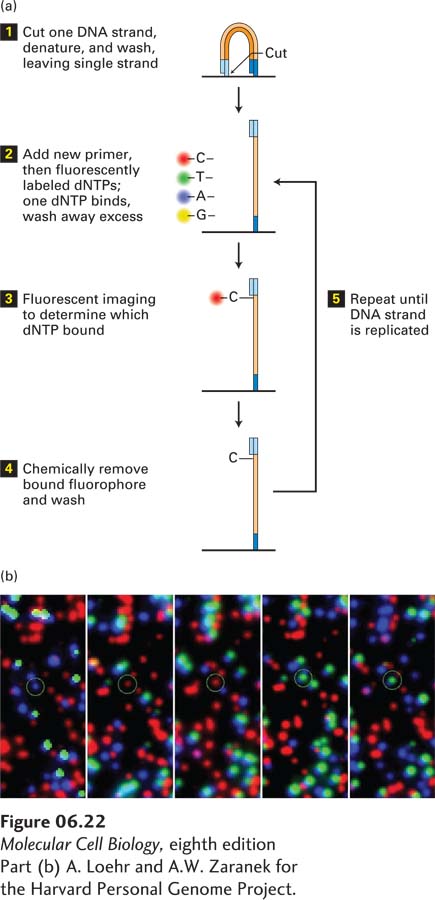
EXPERIMENTAL FIGURE 6- 22 Using fluorescent- tagged deoxyribonucleotide triphosphates for sequence determination. (a) The reaction begins with the cleaving of one strand of the amplified, clustered DNA (see Figure 6- 21 ). After denaturation, a single DNA strand remains attached to the substratum. A synthetic oligodeoxynucleotide is used as the primer for the polymerization reaction, which also contains dNTPs, each fluorescently tagged with a different color. The fluorescent tag is designed to block the 3′ OH group on the dNTP so that once the fluorescent dNTP has been incorporated, further elongation is not possible. Because DNA polymerase will incorporate the same fluorescently labeled dNTP into each of the thousand or so identical DNA copies in a cluster, the entire cluster will be uniformly labeled with the same fluorescent color, which can be imaged in a special microscope. (b) Five images from the same field of view, each corresponding to an individual cycle of dNTP addition. Each colored dot represents a cluster of identical DNA fragments. After each image is made, the fluorescent tags are removed by a chemical reaction that leaves a new primer terminus available for the next cycle of dNTP addition. As can be seen for the circled colored dot, the color changes in each reaction cycle according to which nucleotide is added to the DNA fragments. A typical sequencing reaction may carry out a hundred polymerization cycles, allowing a hundred bases of sequence for each cluster to be determined. Thus a total sequencing reaction of this type may generate as much as 3 × 1011 bases of sequence information in about two days.
[Part (b) A. Loehr and A.W. Zaranek for the Harvard Personal Genome Project.]
[Leave] [Close]