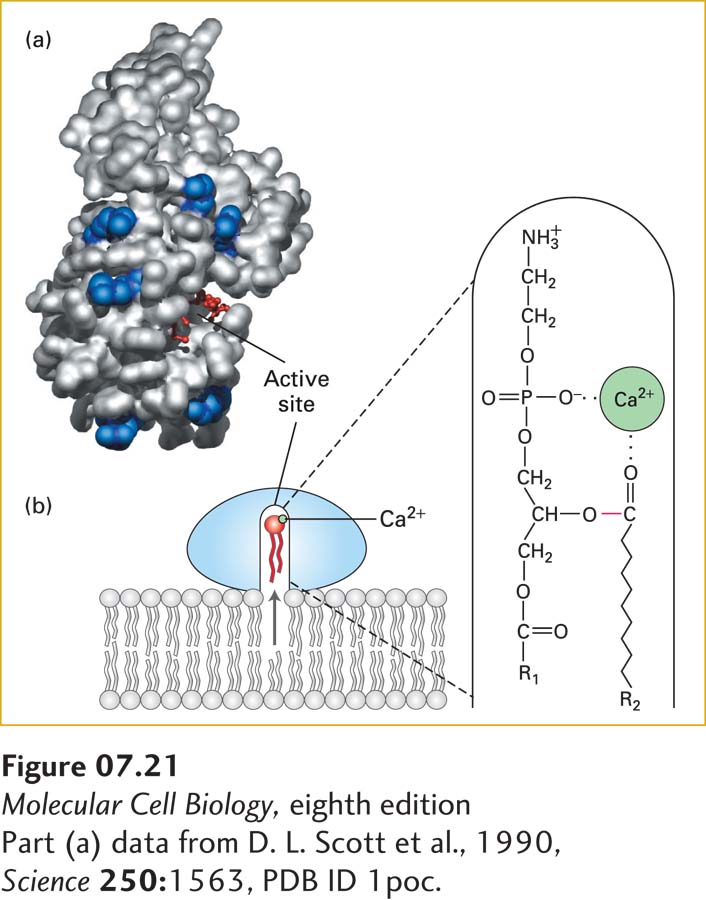
FIGURE 7- d- d- l- d- e-
[Part (a) data from D. L. Scott et al., 1990, Science 250:1563, PDB ID 1poc.]