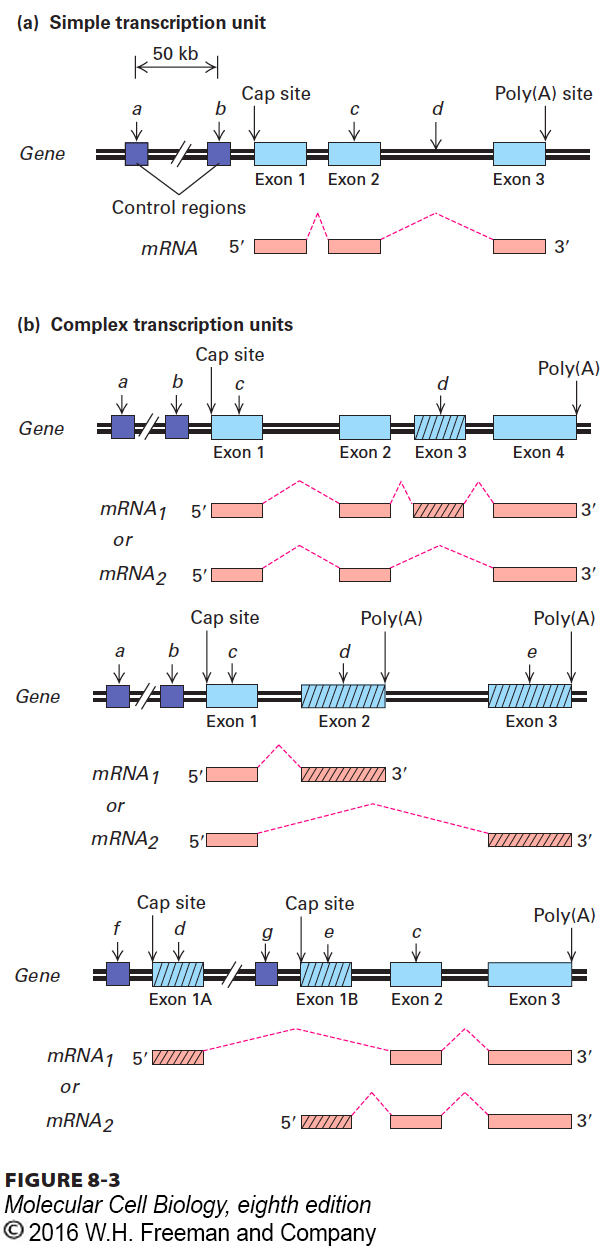
FIGURE 8- 3 Simple and complex eukaryotic transcription units. (a) A simple transcription unit includes a region that encodes one protein, extending from the 5′ cap site to the 3′ poly(A) site, and associated control regions. Introns lie between exons (light blue rectangles) and are removed during processing of the primary transcripts (dashed red lines); thus they do not occur in the functional monocistronic mRNA. Mutations in a transcription- control region (a, b) may reduce or prevent transcription, thus reducing or eliminating synthesis of the encoded protein. A mutation within an exon (c) may result in an abnormal protein with diminished activity. A mutation within an intron (d) that introduces a new splice site results in an abnormally spliced mRNA encoding a nonfunctional protein. (b) Complex transcription units produce primary transcripts that can be processed in alternative ways. (Top) If a primary transcript contains alternative splice sites, it can be processed into mRNAs with the same 5′ and 3′ exons but different internal exons. (Middle) If a primary transcript has two poly(A) sites, it can be processed into mRNAs with alternative 3′ exons. (Bottom) If alternative promoters (f or g) are active in different cell types, mRNA1, produced in a cell type in which f is activated, has a different first exon (1A) than mRNA2, which is produced in a cell type in which g is activated (and in which exon 1B is used). Mutations in control regions (a and b) and in regions within exons shared by the alternative mRNAs (designated c) affect the proteins encoded by both alternatively processed mRNAs. In contrast, mutations (designated d and e) within exons unique to one of the alternatively processed mRNAs affect only the protein translated from that mRNA. For genes that are transcribed from different promoters in different cell types (bottom), mutations in different control regions (f and g) affect expression only in the cell type in which that control region is active.
[Leave] [Close]