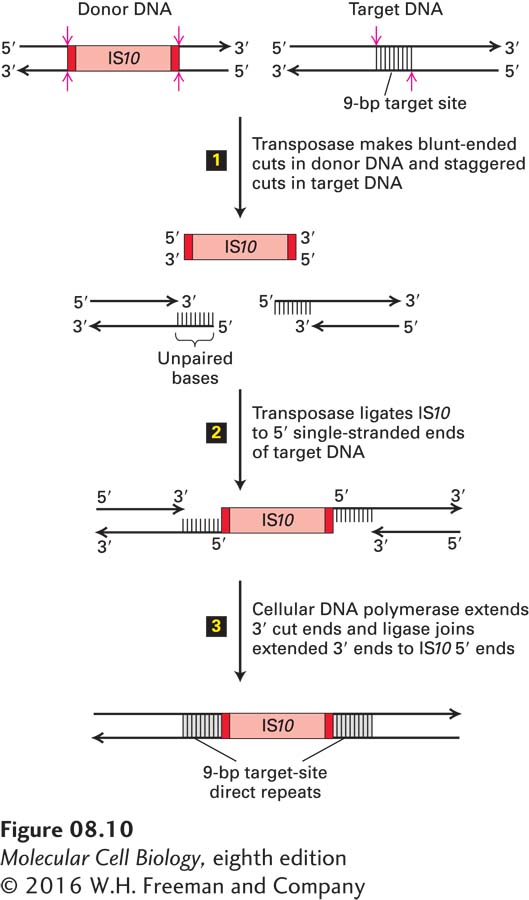
FIGURE 8- 10 Model for transposition of bacterial insertion sequences. Step 1: Transposase, which is encoded by the IS element (IS10 in this example), cleaves both strands of the donor DNA next to the inverted repeats (dark red), excising the IS10 element. At a largely random target site, transposase makes staggered cuts in the target DNA. In the case of IS10, the two cuts are 9 bp apart. Step 2: Ligation of the 3′ ends of the excised IS element to the staggered sites in the target DNA is also catalyzed by transposase. Step 3: The 9- bp gaps of single- stranded DNA left in the resulting intermediate are filled in by a cellular DNA polymerase; finally, cellular DNA ligase forms the 3′→5′ phosphodiester bonds between the 3′ ends of the extended target DNA strands and the 5′ ends of the IS10 strands. This process results in duplication of the target- site sequence on each side of the inserted IS element. Note that the lengths of the target site and IS10 are not to scale. See H. W. Benjamin and N. Kleckner, 1989, Cell 59:373; and 1992, Proc. Natl. Acad. Sci. USA 89:4648.
[Leave] [Close]