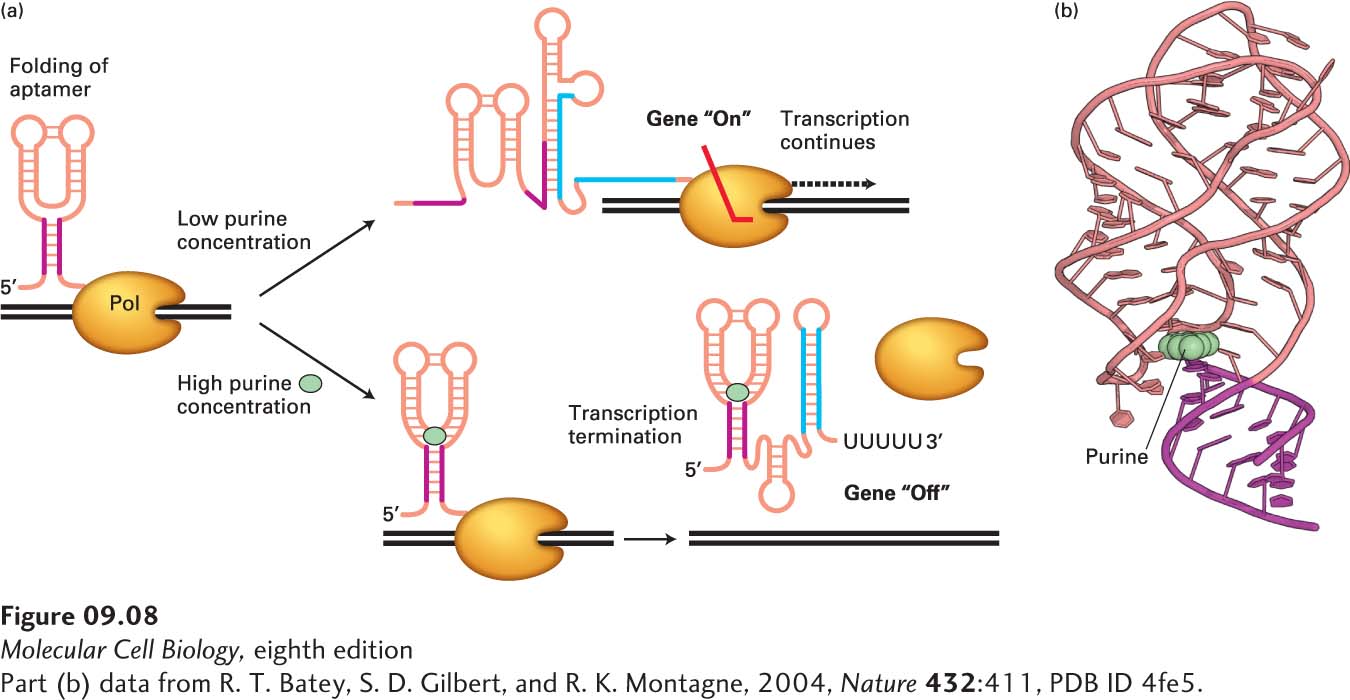
FIGURE 9- 8 Riboswitch control of transcription termination in B. subtilis. (a) During transcription of the Bacillus subtilis xpt- pbuX operon, which encodes enzymes involved in purine synthesis, the 5′ untranslated region of the mRNA can fold into alternative structures depending on the concentration of purines in the cytoplasm, forming the “purine riboswitch.” At high concentrations of purines, the riboswitch folds into an aptamer that binds a purine ligand (cyan circle), allowing formation of a stem- loop transcription termination signal similar to the termination signal that forms in the E. coli trp operon mRNA at high tryptophan concentrations (see Figure 9- 7 ), i.e., a stem loop followed by a run of Us. At low purine concentrations, an alternative RNA structure forms that prevents formation of the transcription termination signal, permitting transcription of the operon. Note the alternative base pairing of the red and blue regions of the RNA. (b) Structure of the purine riboswitch bound to a purine (cyan) as determined by X- ray crystallography. See A. D. Garst, A. L. Edwards, and R. T. Batey, 2011, Cold Spring Harb. Perspect. Biol. 3:a003533.
[Part (b) data from R. T. Batey, S. D. Gilbert, and R. K. Montagne, 2004, Nature 432:411, PDB ID 4fe5.]
[Leave] [Close]