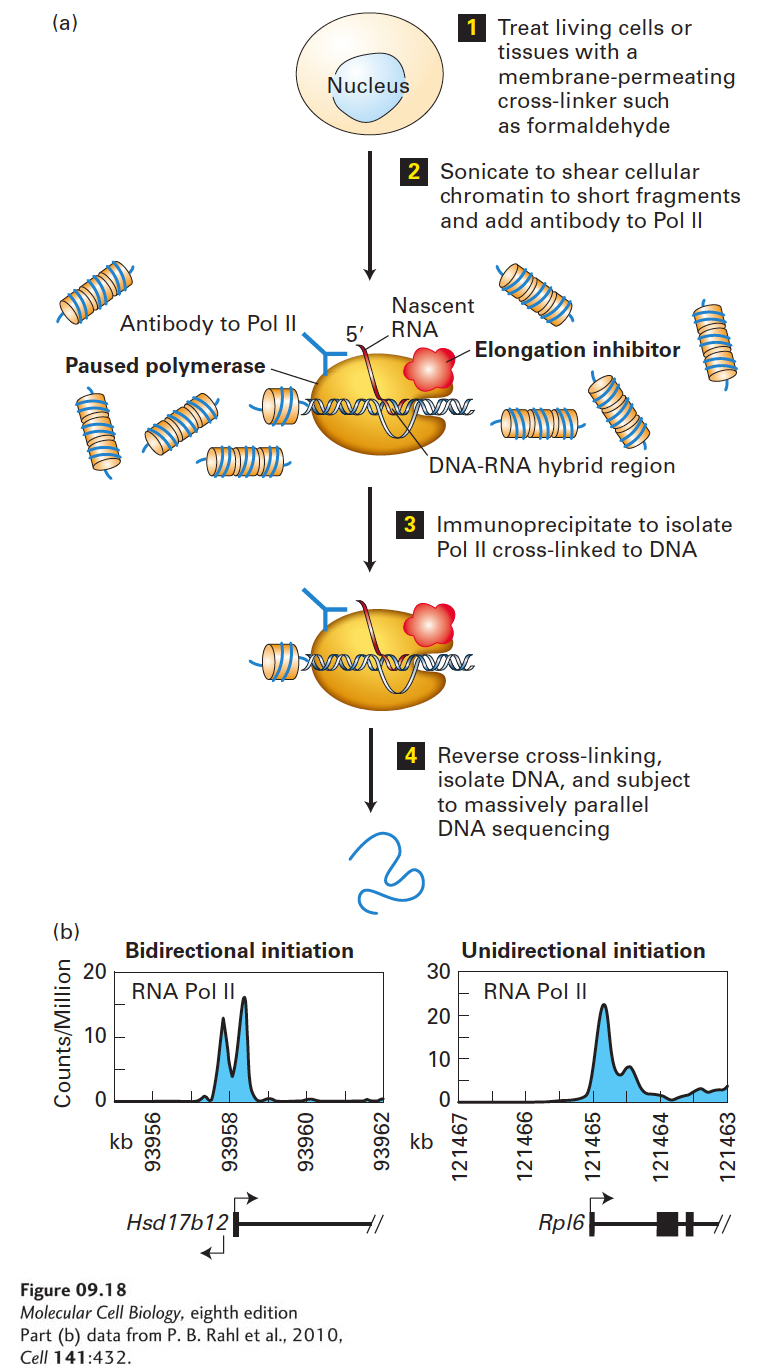
EXPERIMENTAL FIGURE 9- s- 0– s- 6- 0-
[Part (b) data from P. B. Rahl et al., 2010, Cell 141:432.]