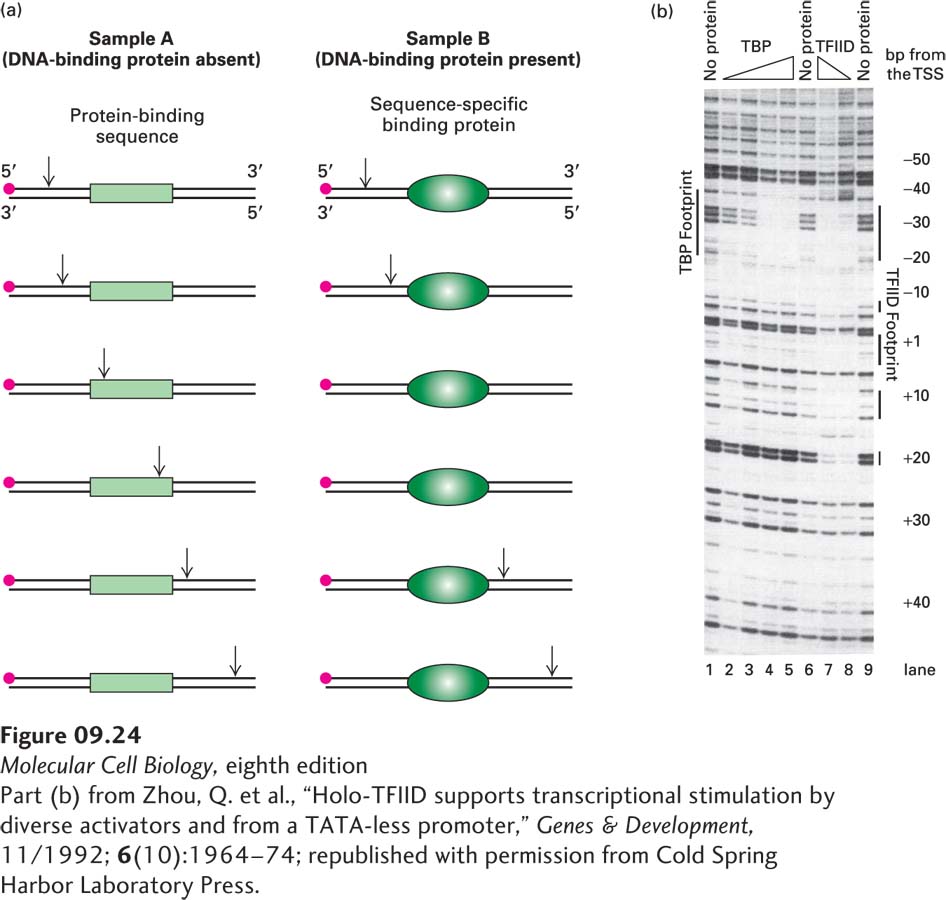
EXPERIMENTAL FIGURE 9- n- e- A- n-
[Part (b) from Zhou, Q. et al., “Holo- A- 4–