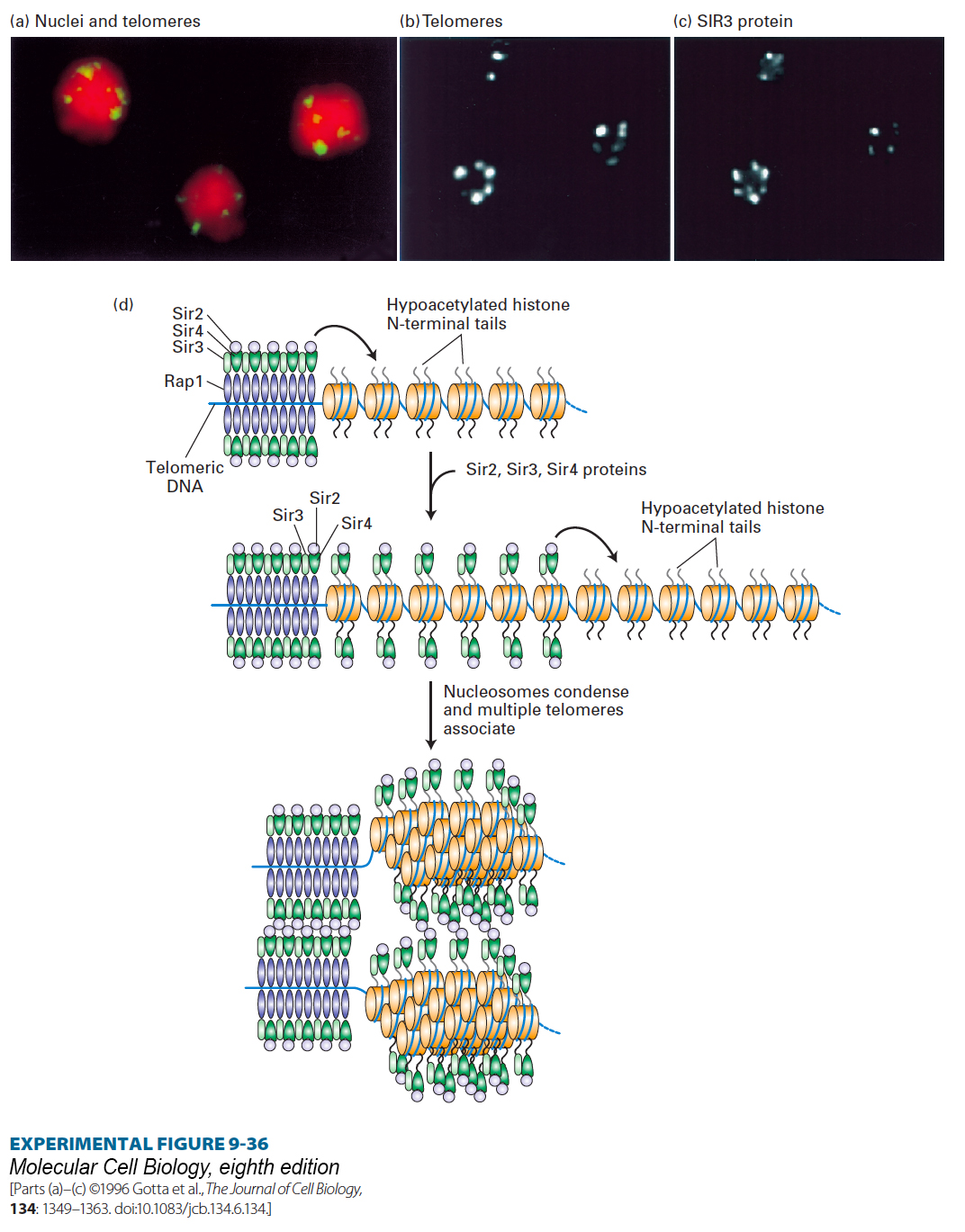
EXPERIMENTAL FIGURE 9- e- e- t- 1- a– r-
[Parts (a)–(c) ©1996 Gotta et al., The Journal of Cell Biology, 134: 1349–