Mitochondrial Genetic Codes Differ from the Standard Nuclear Code
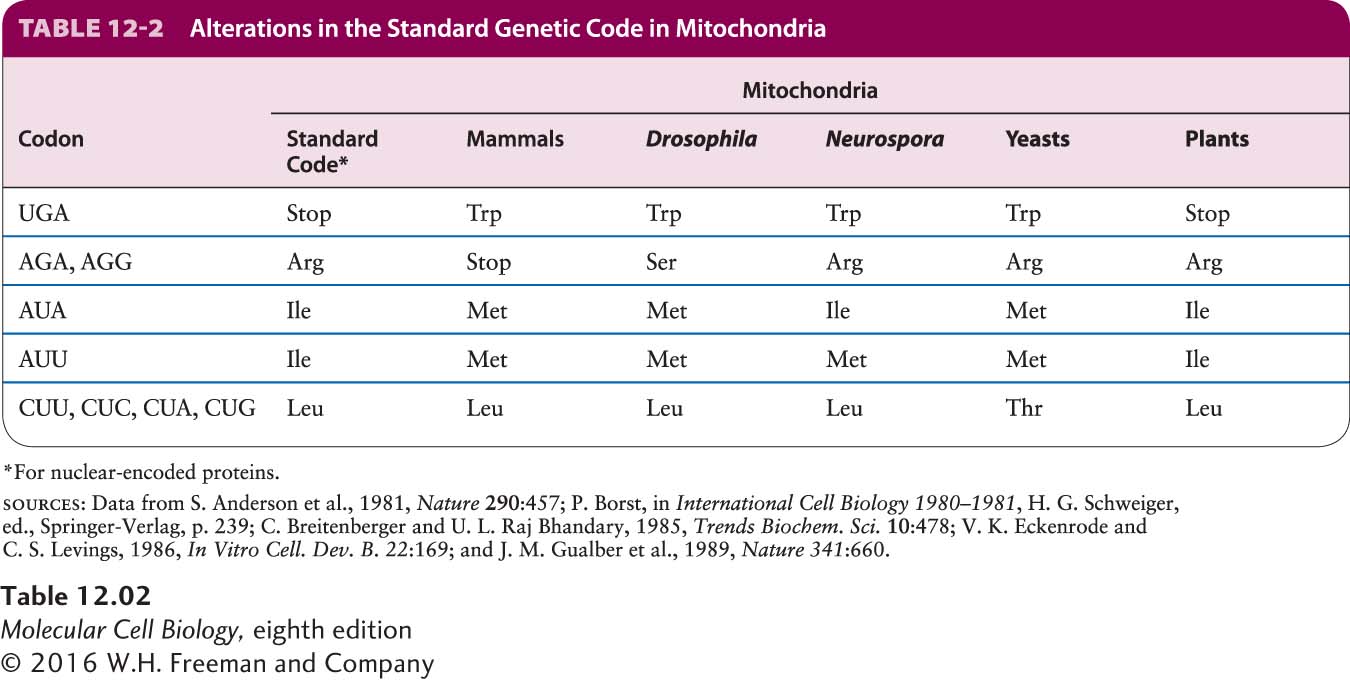
The genetic code used in animal and fungal mitochondria is different from the standard code used in all prokaryotic and eukaryotic nuclear genes; remarkably, the code even differs among mitochondria from different species (Table 12-2). Why and how these differences arose during evolution is a mystery. UGA, for example, is normally a stop codon, but is read as tryptophan by human and fungal mitochondrial translation systems; however, in plant mitochondria, UGA is still recognized as a stop codon. AGA and AGG, the standard nuclear codons for arginine, also code for arginine in fungal and plant mtDNA, but they are stop codons in mammalian mtDNA and serine codons in Drosophila mtDNA.
Page 528
![]() As shown in Table 12-2, plant mitochondria appear to use the standard genetic code. However, comparisons of the amino acid sequences of plant mitochondrial proteins with the nucleotide sequences of plant mtDNAs suggested that CGG could code for either arginine (the “standard” amino acid) or tryptophan. This apparent nonspecificity of the plant mitochondrial code is explained by editing of mitochondrial RNA transcripts, which can convert cytosine residues to uracil residues. If a CGG sequence is edited to UGG, the codon specifies tryptophan, the standard amino acid for UGG, whereas unedited CGG codons encode the standard arginine. Thus the translation system in plant mitochondria does use the standard genetic code.
As shown in Table 12-2, plant mitochondria appear to use the standard genetic code. However, comparisons of the amino acid sequences of plant mitochondrial proteins with the nucleotide sequences of plant mtDNAs suggested that CGG could code for either arginine (the “standard” amino acid) or tryptophan. This apparent nonspecificity of the plant mitochondrial code is explained by editing of mitochondrial RNA transcripts, which can convert cytosine residues to uracil residues. If a CGG sequence is edited to UGG, the codon specifies tryptophan, the standard amino acid for UGG, whereas unedited CGG codons encode the standard arginine. Thus the translation system in plant mitochondria does use the standard genetic code.