DNA Polymerases Introduce Copying Errors and Also Correct Them
The first line of defense in preventing mutations is DNA polymerase itself. Occasionally, when replicative DNA polymerases (Pol δ and Pol ε) progress along the template DNA, an incorrect nucleotide is added to the growing 3′ end of the daughter strand. E. coli DNA polymerases, for instance, introduce about 1 incorrect nucleotide per 104 (ten thousand) polymerized nucleotides. Yet the measured mutation rate in bacterial cells is much lower: about 1 mistake in 109 (one billion) nucleotides incorporated into a growing strand. This remarkable accuracy is largely due to proofreading by E. coli DNA polymerases. Eukaryotic Pol δ and Pol ε employ a similar mechanism.
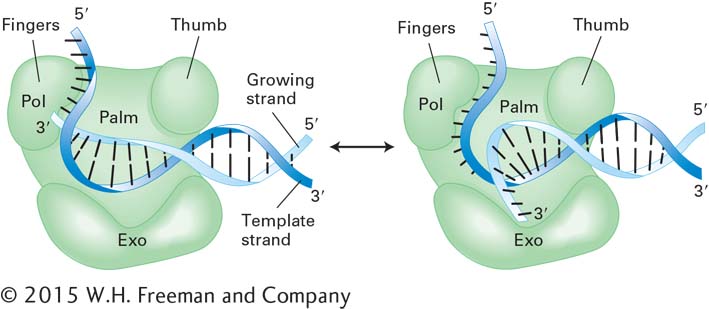
Proofreading depends on the 3′→5′ exonuclease activity of some DNA polymerases. When an incorrect base is incorporated during DNA synthesis, base pairing between the 3′ nucleotide of the nascent strand and the template strand does not occur. As a result, the polymerase pauses, then transfers the 3′ end of the growing chain to its exonuclease site, where the incorrect mispaired base is removed (Figure 5-33). Then the 3′ end is transferred back to the polymerase site, where this region is copied correctly. All three E. coli DNA polymerases have proofreading activity, as do the two eukaryotic DNA polymerases, δ and ε, used for replication of most chromosomal DNA in animal cells. It seems likely that proofreading is indispensable for all cells to avoid excessive mutations.