Viruses Can Be Cloned and Counted in Plaque Assays
The number of infectious viral particles in a sample can be quantified by a plaque assay. This assay is performed by culturing a dilute sample of viral particles on a plate covered with host cells and then counting the number of local lesions, called plaques, that develop (Figure 5-44). A plaque develops on the plate wherever a single virion initially infects a single cell. The virus replicates in this initial host cell and then lyses (ruptures) the cell, releasing many progeny virions that infect the neighboring cells on the plate. After a few such cycles of infection, enough cells are lysed to produce a visible clear area, or plaque, in the layer of remaining uninfected cells. Since all the progeny virions in a plaque are derived from a single parent virus, they constitute a viral clone.
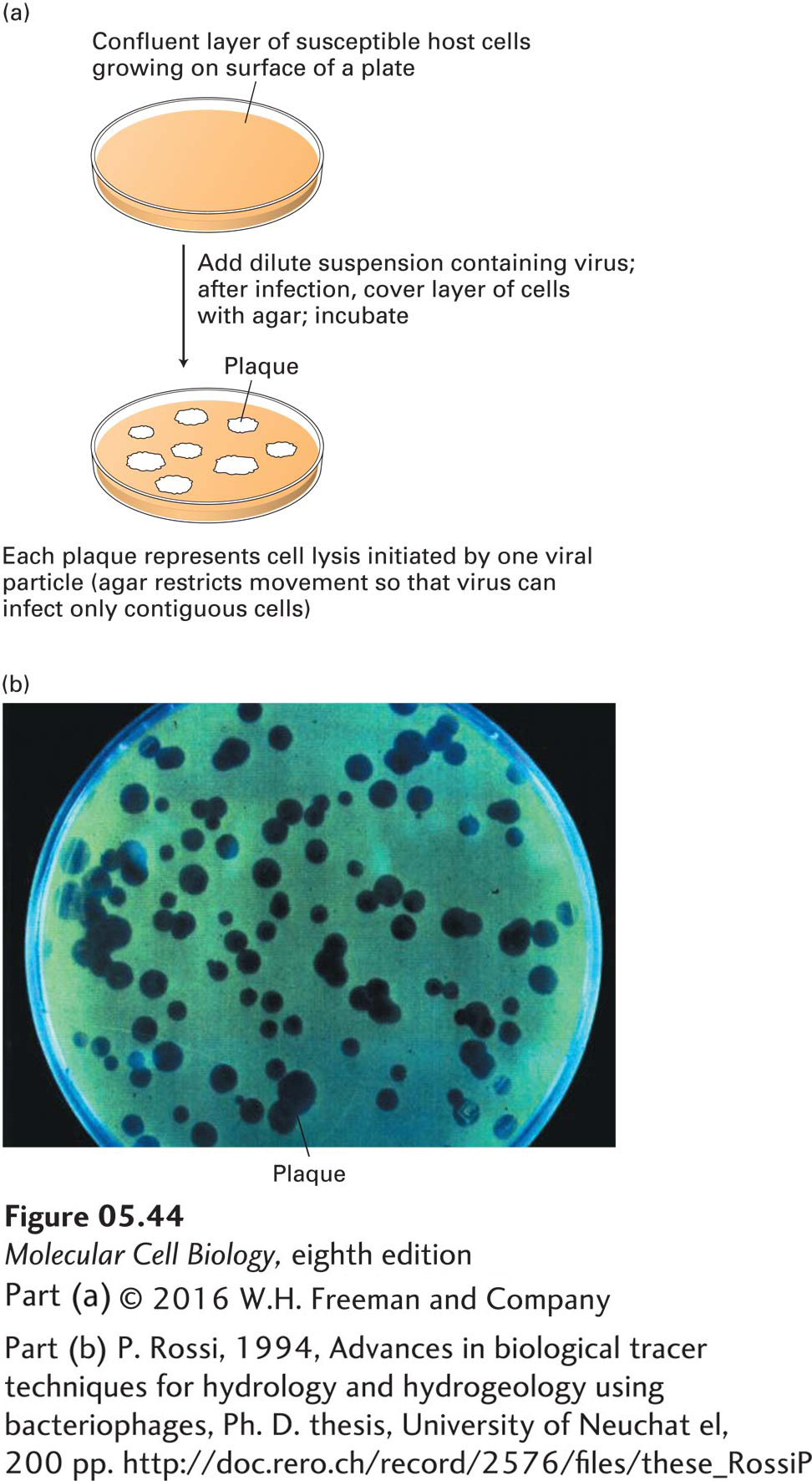
This type of plaque assay is in standard use for bacterial and animal viruses. Plant viruses can be assayed similarly by counting local lesions on plant leaves inoculated with viruses. Analysis of viral mutants, which are commonly isolated by plaque assays, has contributed extensively to our current understanding of molecular cellular processes.