Review the Concepts
1. What are Watson-
2. Preparing plasmid (double-
3. What difference between RNA and DNA helps to explain the greater stability of DNA? What implications does this have for the function of DNA?
4. What are the major differences in the synthesis and structure of prokaryotic and eukaryotic mRNAs?
5. While investigating the function of a specific growth factor receptor gene from humans, researchers found that two types of proteins are synthesized from this gene. A larger protein containing a membrane-
6. The transcription of many bacterial genes relies on functional groups called operons, such as the tryptophan operon (see Figure 5-13a). What is an operon? What advantages are there to having genes arranged in an operon, compared with the arrangement in eukaryotes?
7. How would a mutation in the poly(A)-binding protein gene affect translation? How would an electron micrograph of polyribosomes from such a mutant differ from the normal pattern?
8. What characteristic of DNA results in the requirement that some DNA synthesis be discontinuous? How are Okazaki fragments and DNA ligase used by the cell?
9. Eukaryotes have repair systems that prevent mutations due to copying errors and exposure to mutagens. What are the three excision-
10. DNA repair systems are responsible for maintaining genomic fidelity in normal cells despite the high frequency with which mutational events occur. What type of DNA mutation is generated by (a) UV radiation and (b) ionizing radiation? Describe the system responsible for repairing each of these types of mutations in mammalian cells. Postulate why a loss of function in one or more DNA repair systems typifies many cancers.
11. What is the name given to the process that can repair DNA damage and generate genetic diversity? Briefly describe the similarities and differences of the two processes.
12. The genome of a retrovirus can integrate into the host-
13. a. Which of the following DNA strands, the top or bottom, would serve as a template for RNA transcription if the DNA molecule were to unwind in the indicated direction?
5′ ACGGACTGTACCGCTGAAGTCATGGACGCTCGA 3′
3′ TGCCTGACATGGCGACTTCAGTACCTGCGAGCT 5′

Direction of DNA unwinding
b. What would be the resulting RNA sequence (written 5′→3′ )?
14. Contrast prokaryotic and eukaryotic gene characteristics.
15. You have learned about the events surrounding DNA replication and the central dogma. Identify the steps associated with these processes that would be adversely affected in the following scenarios.
Helicases unwind the DNA, but stabilizing proteins are mutated and cannot bind to the DNA.
The mRNA molecule forms a hairpin loop on itself via complementary base pairing in an area spanning the AUG start site.
The cell is unable to produce functional tRNAiMet.
Page 220
16. Use the key provided below to determine the amino acid sequence of the polypeptide produced from the following DNA sequence. Intron sequences are highlighted. Note: Not all amino acids in the key will be used.
5′ TTCTAAACGCATGAAGCACCGTCTCAGAGCCAGTGA 3′
3′ AAGATTTGCGTACTTCGTGGCAGAGTCTCGGTCACT 5′

Direction of DNA unwinding
Asn = AAU Cys = TCG Gly = CAG His = CAU Lys = AAG
Met = AUG Phe = UUC Ser = AGC Tyr = UAC Val = GUC; GUA
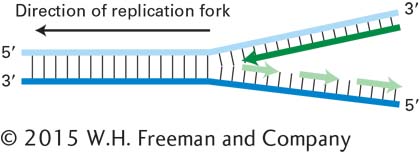
17. a. Look at the figure above. Explain why it is necessary for Okazaki fragments to be formed as the lagging strand is produced (instead of a continuous strand).
b. If the DNA polymerase in the figure above could bind only to the lower template strand, under what condition(s) would it be able to produce a leading strand?
18. The DNA repair systems preferentially target the newly synthesized strand. Why is this important?
19. Identify the specific types of point mutations below (you are viewing the direct DNA version of the RNA sequence).
Original sequence: 5′ AUG TCA GGA CGT CAC TCA GCT 3′
Mutation A: 5′ AUG TCA GGA CGT CAC TGA GCT 3′
Mutation B: 5′ AUA TCA GGA CGT CAC TCA GCT 3′
20. a. Detail the key differences between lytic and nonlytic viral infection and provide an example of each.
b. Which of the following processes occurs in both lytic and nonlytic viral infections?
Infected cell ruptures to release viral particles.
Viral mRNAs are transcribed by the host-
cell translation machinery. Viral proteins and nucleic acids are packaged to produce virions.