Hybridization Techniques Permit Detection of Specific DNA Fragments and mRNAs
Hybridization depends on the ability of complementary single-
Two very sensitive methods for detecting a particular DNA or RNA sequence within a complex mixture combine separation by gel electrophoresis and hybridization to a complementary DNA probe that is either radioactively or fluorescently labeled. A third method involves hybridizing labeled probes directly to a prepared tissue sample. We will encounter references to all three of these techniques, which have numerous applications, in other chapters.
Southern Blotting The first hybridization technique developed to detect DNA fragments of a specific sequence is known as Southern blotting, after its originator, E. M. Southern. This technique is capable of detecting a single specific restriction fragment in the highly complex mixture of fragments produced by cleavage of the entire human genome with a restriction enzyme. A common procedure for separating DNA fragments of different sizes is gel electrophoresis. Near neutral pH, DNA molecules carry a large negative charge and therefore move toward the positive electrode during gel electrophoresis. Because the gel matrix restricts random diffusion, molecules of the same length migrate together as distinct bands. Smaller molecules move through the gel matrix more readily than larger molecules. Larger DNA molecules from about 200 bp to more than 20 kb can be separated electrophoretically on agarose gels, and smaller DNA molecules from about 10 to 2000 bp can be separated on polyacrylamide gels.
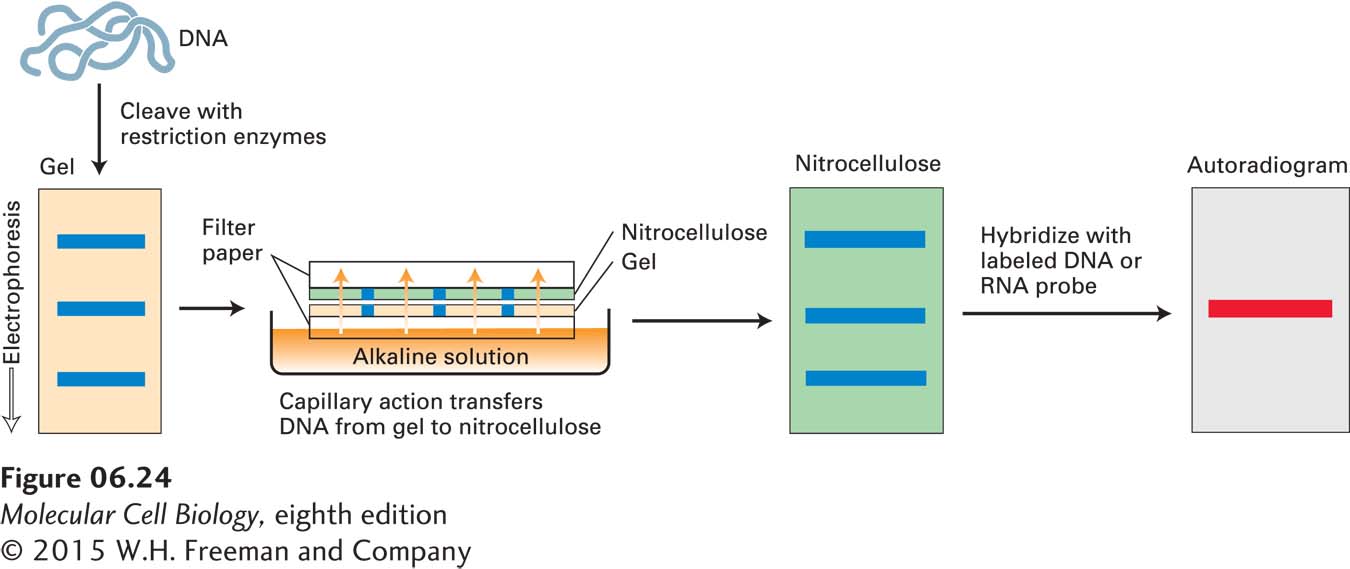
When DNA from a whole genome is fragmented by digestion with a restriction enzyme, the mixture of DNA fragments that is produced is so complex that even after gel electrophoresis, so many different fragments of nearly the same length are present that it is not possible to resolve any particular DNA fragment as a discrete band on the gel. Nevertheless, it is possible to identify a particular fragment migrating as a band on the gel by its ability to hybridize to a specific DNA probe. To accomplish this, the restriction fragments present in the gel are denatured with alkali and transferred onto a nitrocellulose filter or nylon membrane by blotting (Figure 6-24). This procedure preserves the distribution of the fragments in the gel, creating a replica of the gel on the filter. (The blot is used because probes do not readily diffuse into the original gel.) The filter is then incubated under hybridization conditions with a specific labeled DNA probe, which is usually generated from a cloned restriction fragment. The DNA restriction fragment that is complementary to the probe hybridizes to it, and its location on the filter can be revealed by autoradiography (for a radiolabeled probe) or by fluorescent imaging (for a fluorescently labeled probe). Although PCR is now more commonly used to detect the presence of a particular sequence in a complex mixture, Southern blotting is still useful for reconstructing the relationship between genomic sequences that are too far apart to be amplified by PCR in a single reaction.
Page 247
Northern Blotting One of the most basic ways to characterize a cloned gene is to determine when and where in an organism the gene is expressed. The expression of a particular gene can be followed by assaying for the corresponding mRNA by Northern blotting, named, in a play on words, after the related method of Southern blotting. An RNA sample, often the total cellular RNA, is denatured by treatment with an agent such as formaldehyde that disrupts the hydrogen bonds between base pairs, ensuring that all the RNA molecules have an unfolded, linear conformation. The individual RNAs are separated according to size by gel electrophoresis and transferred to a nitrocellulose filter to which the extended denatured RNAs adhere. As in Southern blotting, the filter is then exposed to a labeled DNA probe that is complementary to the gene of interest; finally, the labeled filter is subjected to autoradiography. Because the amount of a specific RNA in a sample can be estimated from a Northern blot, the procedure is widely used to compare the amounts of a particular mRNA in cells under different conditions.
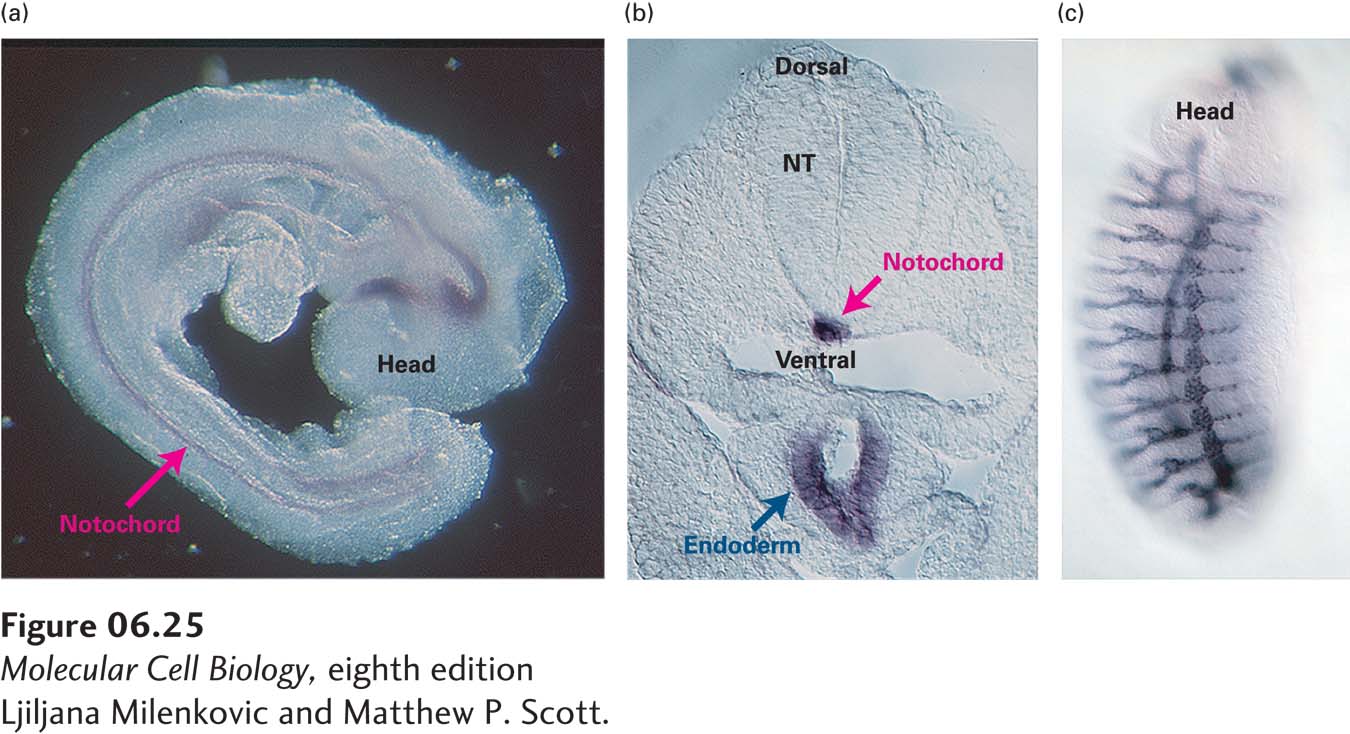
In Situ Hybridization Northern blotting requires extracting the mRNA from a cell or mixture of cells, which means that the cells are removed from their normal location within an organism or tissue. As a result, the location of a cell and its relation to its neighbors is lost. To retain such positional information in precise studies of gene expression, a whole or sectioned tissue, or even a whole permeabilized embryo, may be subjected to in situ hybridization to detect the mRNA encoded by a particular gene. This technique allows gene transcription to be monitored in both time and space (Figure 6-25).