Chapter Introduction
CHAPTER 8: Genes, Genomics, and Chromosomes
In previous chapters, we learned how the structure and composition of proteins allow them to perform a wide variety of cellular functions. We also examined another vital component of cells, the nucleic acids, and the process by which information encoded in the sequence of DNA is translated into protein. In this chapter, our focus again is on DNA and proteins as we consider the characteristics of eukaryotic nuclear genomes: the features of genes and the other DNA sequences that constitute the genome, and how this DNA is structured and organized by proteins within the cell.
By the beginning of the twenty-
Surprisingly, DNA sequencing revealed that large portions of the genomes of metazoans and plants do not encode mRNAs or any other RNAs required by the organism. Remarkably, such noncoding DNA constitutes about 98.5 percent of human chromosomal DNA! The noncoding DNA in multicellular organisms contains many regions that are similar, but not identical, to one another. Variations within some stretches of this repetitious DNA between individuals are so great that every person can be distinguished by a DNA “fingerprint” based on these sequence variations. Moreover, some repetitious DNA sequences are not found in the same positions in the genomes of different individuals of the same species. At one time, all noncoding DNA was collectively termed “junk DNA” and was considered to serve no purpose. We now understand the evolutionary basis of all this extra DNA, and of the variation in location of certain sequences between individuals. Cellular genomes harbor transposable (mobile) DNA elements that can copy themselves and move throughout the genome. Although transposable DNA elements seem to have little function in the life cycle of an individual organism, over evolutionary time they have shaped our genomes and contributed to the rapid evolution of multicellular organisms. Moreover, we now understand that much of the DNA that does not encode proteins or stable RNAs functions as binding sites for protein complexes that regulate gene transcription.
Page 302
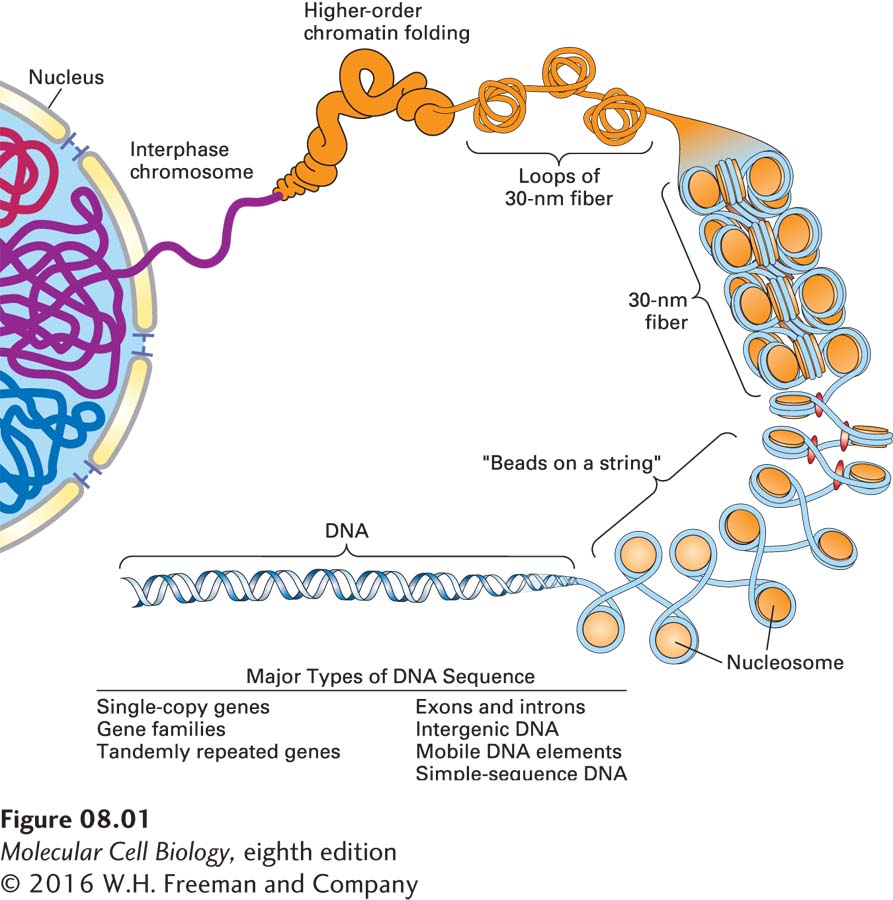
The sheer length of cellular DNA is a significant problem with which cells must contend. The DNA in a single human cell, which measures about 2 m in total length, must be contained within nuclei with diameters of less than 10 µm, a compaction ratio of greater than 105 to 1. In relative terms, if a cell were 1 cm in diameter (about the size of a pea), the length of DNA packed into its nucleus would be about 2 km (1.2 miles)! Specialized eukaryotic proteins associated with nuclear DNA exquisitely fold and organize the DNA so that it fits into nuclei. And yet, at the same time, any given portion of this highly compacted DNA can be accessed readily for transcription, replication, and repair of damage without the long DNA molecules becoming tangled or broken. Furthermore, the integrity of DNA must be maintained during the process of cell division when it is partitioned into daughter cells. In eukaryotes, the complex of DNA and the proteins that organize it, called chromatin, can be visualized as individual chromosomes during mitosis. As we will see in this and the following chapter, the organization of DNA into chromatin allows a mechanism for regulation of gene expression that is not available in bacteria.
In the first four sections of this chapter, we provide an overview of the landscape of eukaryotic genes and genomes. First we discuss the structure of eukaryotic genes and the complexities that arise in higher organisms from the processing of mRNA precursors into alternatively spliced mRNAs. Next we discuss the main classes of eukaryotic DNA, including the special properties of transposable DNA elements and how they have shaped contemporary genomes. This background prepares us to discuss genomics, computer-