
Chapter 6. Complementation in Yeast
Introduction

Analyze the Data 6-1: Complementation in Yeast
A culture of yeast that requires uracil for growth (ura3−) was treated with a mutagen, and two mutant colonies, X and Y, were isolated. Mating type a cells of mutant X were mated with mating type α cells of mutant Y to form diploid cells. The parental (ura3−), X, Y, and diploid cells were all streaked onto agar plates containing uracil and incubated at 23 °C or 32 °C. Cell growth was monitored by the formation of colonies on the plates, as shown in the figure below.
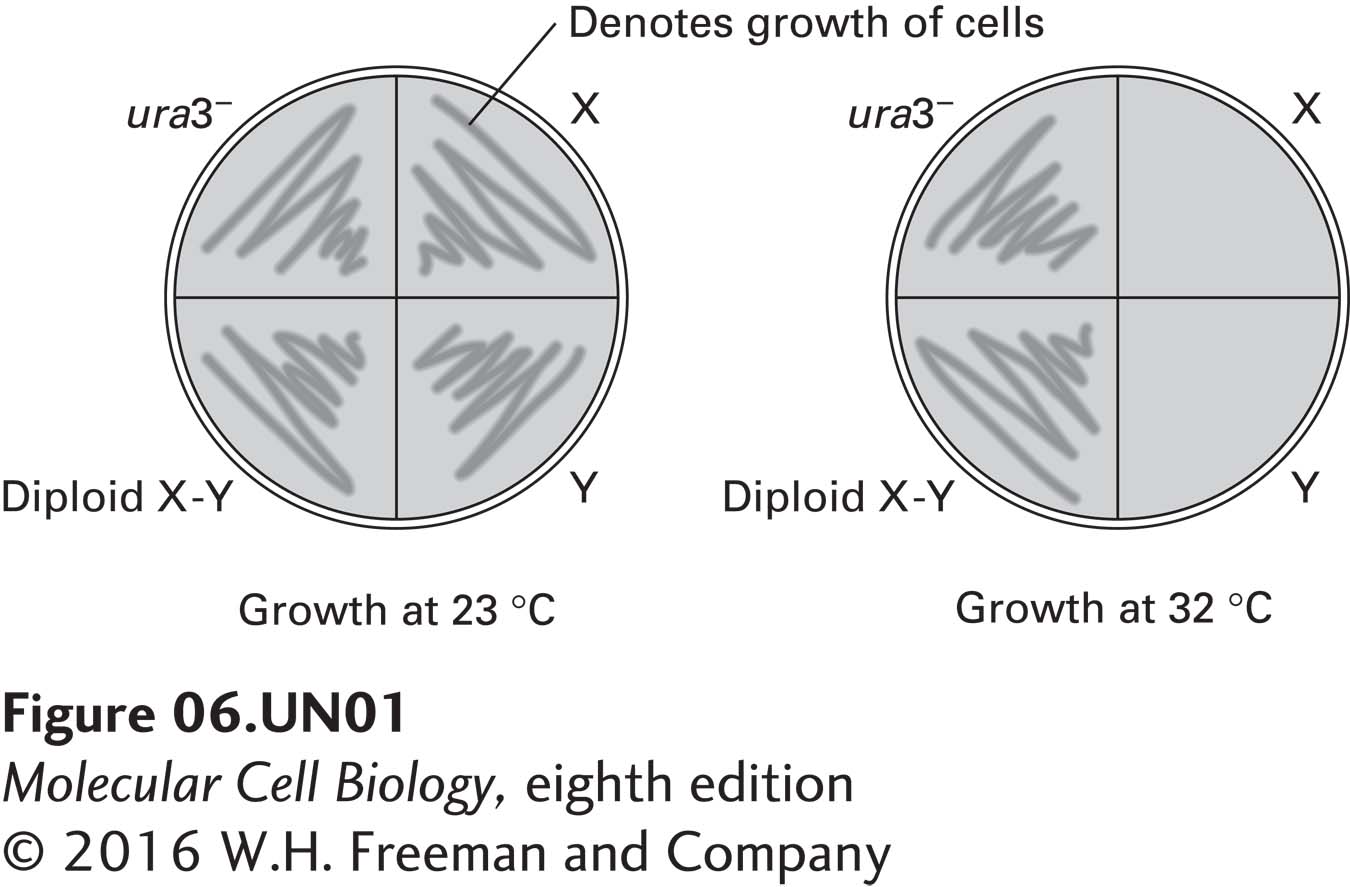
a. What can be deduced about mutants X and Y from the data provided?
b. A wild-type yeast cDNA library, prepared in a plasmid that contains the wild-type URA3+ gene, was used to transform X cells, which were then cultured as indicated. Each black spot below represents a single clone growing on a petri plate. What are the molecular differences between the clones growing on the two plates? How can these results be used to identify the plasmid that contains a wild-type copy of gene X? Based on these results, how can the identity of gene X be uncovered?
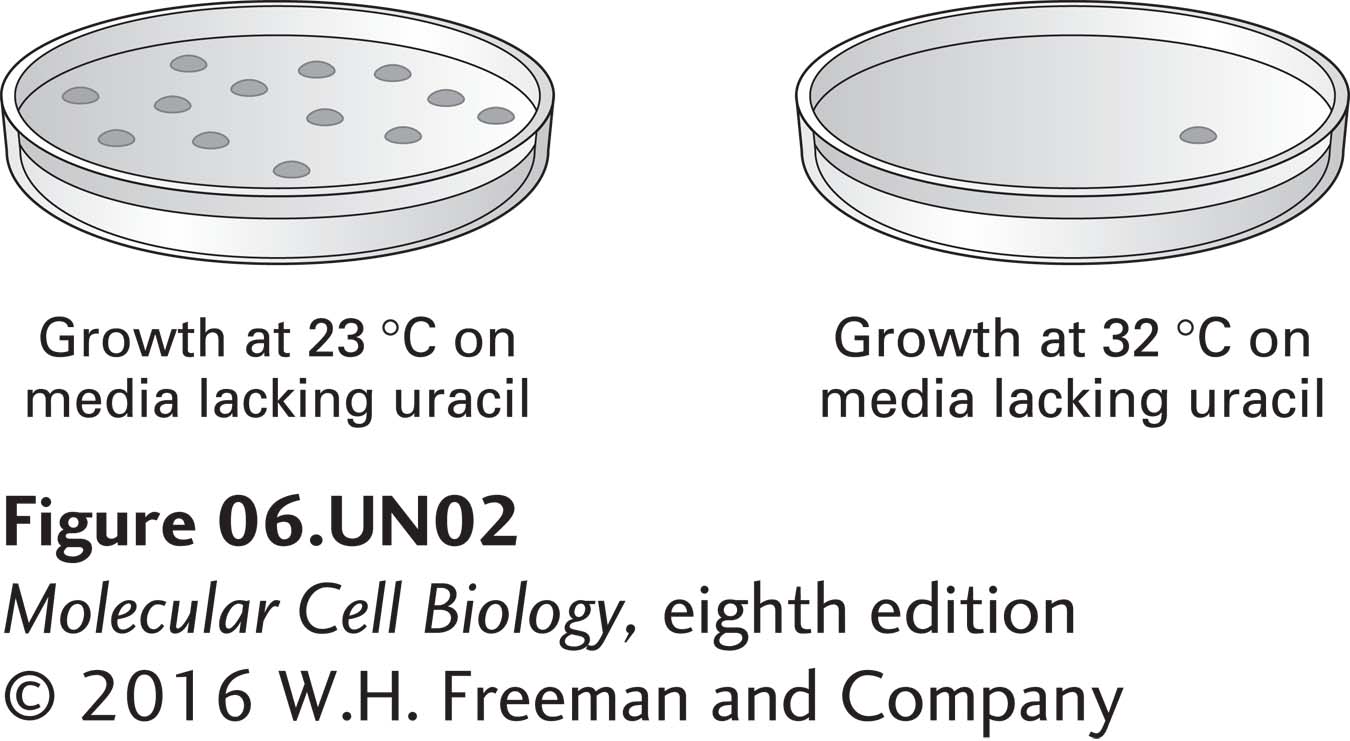
The plate on the left lacks uracil; clones growing on this plate must be able to synthesize their own uracil (they must have a wild-type copy of the URA3 gene). The yeast themselves are defective in uracil synthesis, so each clone on the left plate must be transformed with a plasmid containing the URA3 gene. The single clone on the plate at the right grew in the absence of uracil at the restrictive temperature. Therefore, it must harbor a plasmid that contains the wild-type copy of the X gene, allowing mutant X cells to grow at 32°C. If the plasmid were to be re-isolated from this clone and the yeast cDNA insert analyzed and sequenced, the X gene will have been identified. Extract the plasmid from the complemented yeast and sequence it. The sequencing primer will be complementary to the vector backbone.
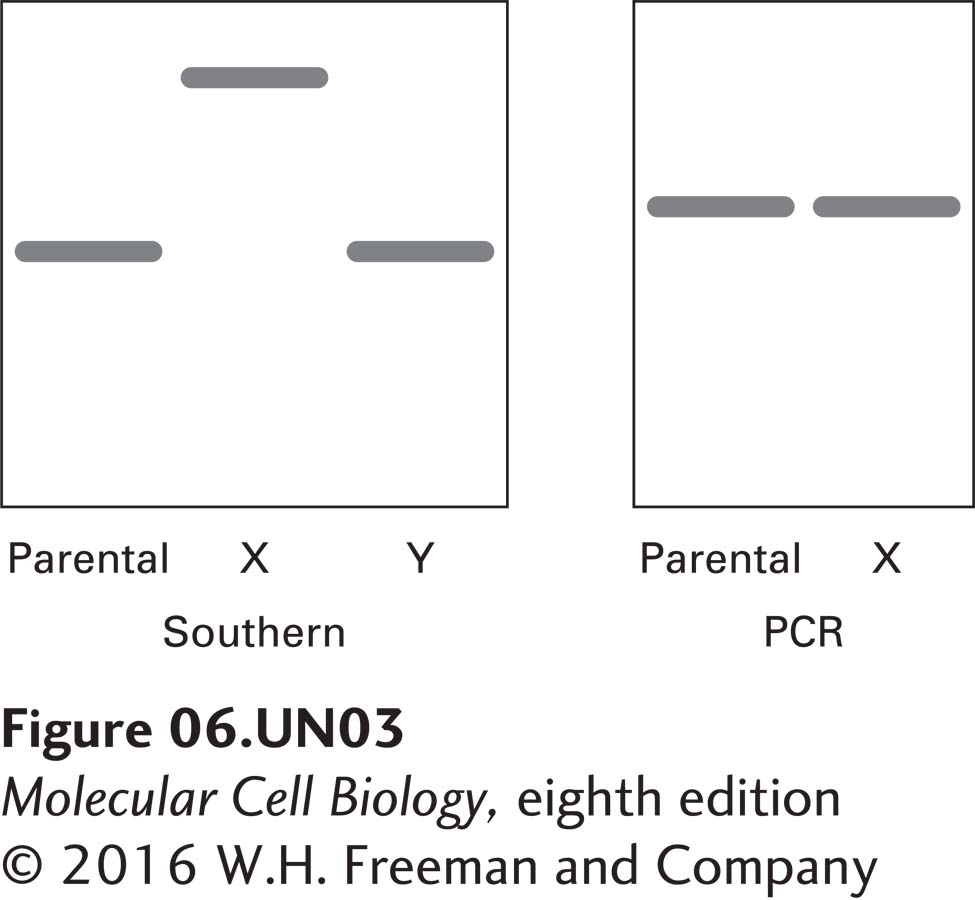
c. DNA was extracted from the parent cells, from X cells, and from Y cells. PCR primers were used to amplify the gene encoding protein X in both the parent and the X cells. The primers were complementary to regions of DNA just external to the gene encoding X. The PCR results are shown in the gel at the right. What can be deduced about the mutation in the X gene from these data?
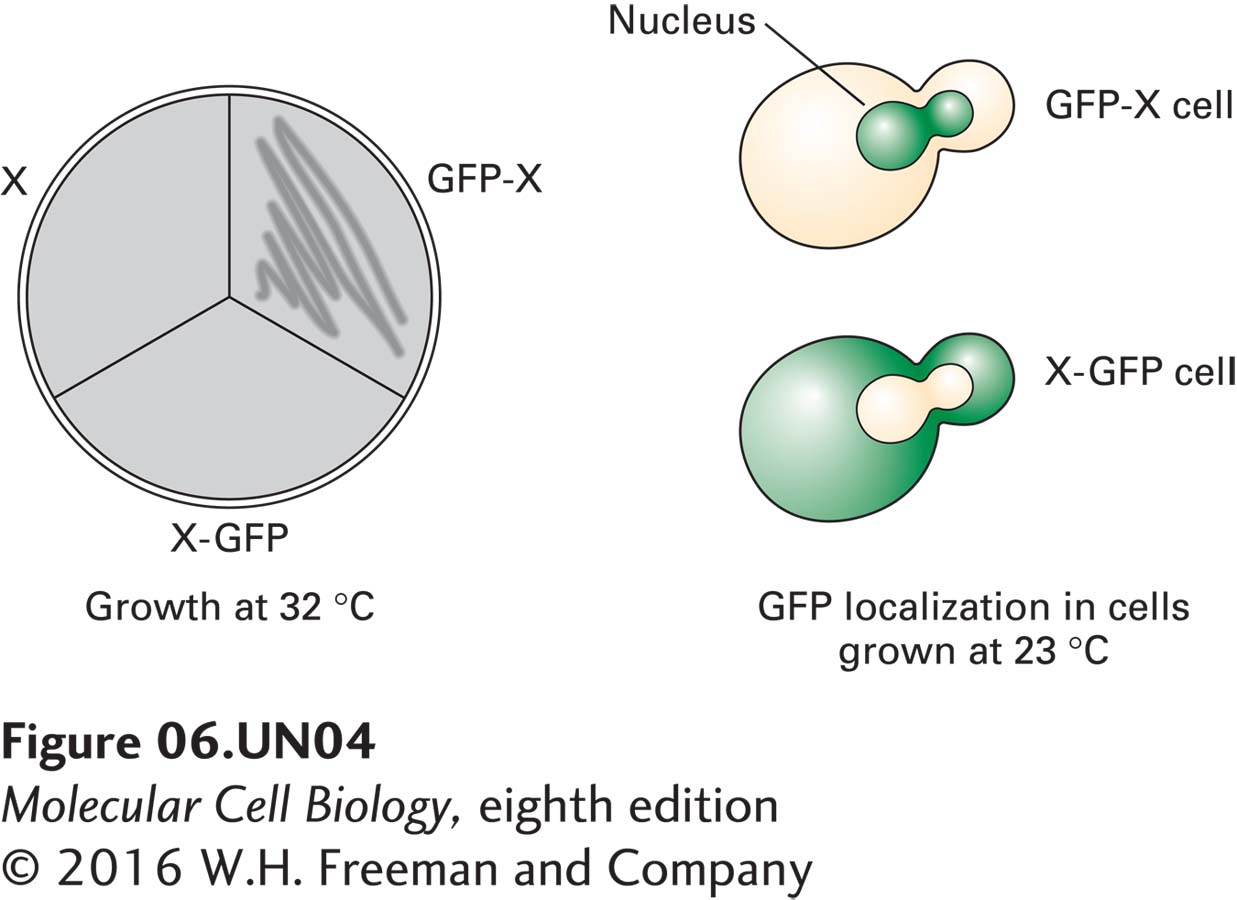
d. Constructs of the wild-type gene X were engineered to encode fusion proteins in which green fluorescent protein (GFP) is present at the N-terminus (GFP-X) or the C-terminus (X-GFP) of protein X. Both constructs, present on a URA3+ plasmid, were used to transform X cells grown in the absence of uracil. The transformants were then monitored for growth at 32 °C, as shown below at the left. At the right are typical fluorescent images of X-GFP and GFP-X cells grown at 23 °C, in which green denotes the presence of green fluorescent protein. What is a reasonable explanation for the growth of GFP-X but not X-GFP cells at 32 °C?
e. Haploid offspring of the diploid cells from part (a) above were generated. XY double mutants constituted ¼ of these offspring. Haploid X cells, Y cells, and XY cells in liquid culture were synchronized at a stage just prior to budding and then shifted from 23 °C to 32 °C. Examination of the cells 24 hours later revealed that X cells were arrested with small buds, Y cells were arrested with large buds, and XY cells were arrested with small buds. What is the relationship between X and Y?
Activity results are being submitted...