
Chapter 9. Coordination of Eukaryotic Polymerases
Introduction

Analyze the Data 9-1: Coordination of Eukaryotic Polymerases
In eukaryotes, the three RNA polymerases, Pol I, II, and III, each transcribe unique genes required for the synthesis of ribosomes: 25S and 18S rRNAs (Pol I), 5S rRNA (Pol III), and mRNAs for ribosomal proteins (Pol II). Researchers have long speculated that the activities of the three RNA polymerases are coordinately regulated according to the demand for ribosome synthesis: high demand in replicating cells in rich nutrient conditions and low demand when nutrients are scarce. To determine whether the activities of the three polymerases are coordinated, Arnaud Laferte and colleagues engineered a strain of yeast to be partially resistant to the inhibition of cell growth by the drug rapamycin (2006, Genes Dev. 20:2030–2040). As discussed in Chapter 10, rapamycin inhibits a protein kinase called TOR (for target of rapamycin) that regulates the overall rate of protein synthesis and ribosome synthesis. When TOR is inhibited by rapamycin, the transcription of rRNAs by Pol I and Pol III and of ribosomal protein mRNAs by RNA polymerase II are all rapidly repressed. Part of the inhibition of rRNA synthesis results from the dissociation of the Pol I transcription factor Rrn3 from Pol I. In the strain constructed by Laferte and colleagues, the wild-type Rrn3 gene and the wild-type A43 gene, encoding the Pol I subunit to which Rrn3 binds, were replaced with a gene encoding a fusion of the A43 Pol I subunit with Rrn3. The idea was that the covalent fusion of the two proteins would prevent the Rrn3 dissociation from Pol I otherwise caused by rapamycin treatment. The resulting CARA (constitutive association of Rrn3 and A43) yeast strain was found to be partially resistant to rapamycin. In the absence of rapamycin, the CARA strain grew at the same rate and had as many ribosomes as wild-type cells.
a. To analyze rRNA transcription by Pol I, total RNA was isolated from rapidly growing wild-type (WT) cells and from CARA cells at various times following the addition of rapamycin. The concentration of the 35S rRNA precursor transcribed by Pol I (see Figure 10-41) was assayed by the primer-extension method. Since the 5′ end of the 35S rRNA precursor is degraded during the processing of 28S and 18S rRNA, this method measures the relatively shortlived pre-rRNA precursor. It is therefore an indirect measure of the rate of rRNA transcription by Pol I. The results of this primer-extension assay are shown below. How does the CARA strain’s Pol I–Rrn3 fusion affect the response of Pol I transcription to rapamycin?
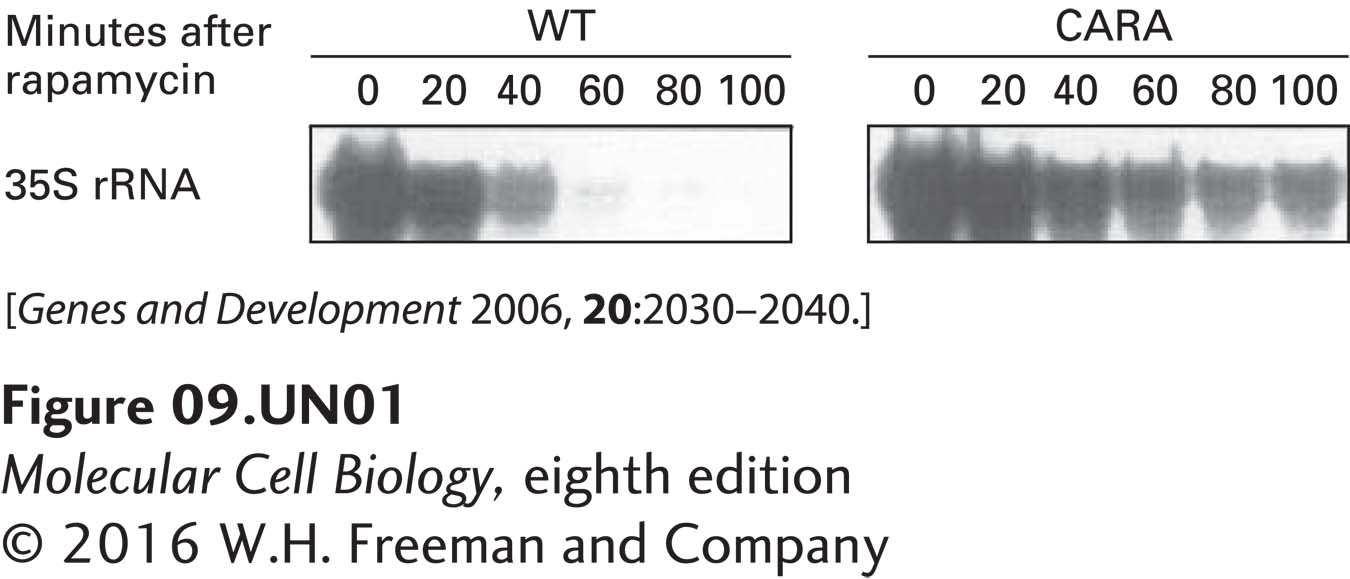
In untreated cells (t = 0 minutes), both wild-type and CARA cells transcribed equivalent amounts of ribosomal RNAs, suggesting that the mutant Pol I functions normally under control conditions. This finding is consistent with the observation that the growth rate of yeast harboring this mutation was indistinguishable from wild-type cells. However, when the drug rapamycin was added, Pol I transcription in wild-type cells was completely inhibited by 100 minutes after addition of the drug. In contrast, in the CARA cells, Pol I transcription was much less inhibited and continued at a significant level even at 100 minutes after addition of rapamycin. Consequently, Pol I transcription in the CARA cells was only partially inhibited by rapamycin.
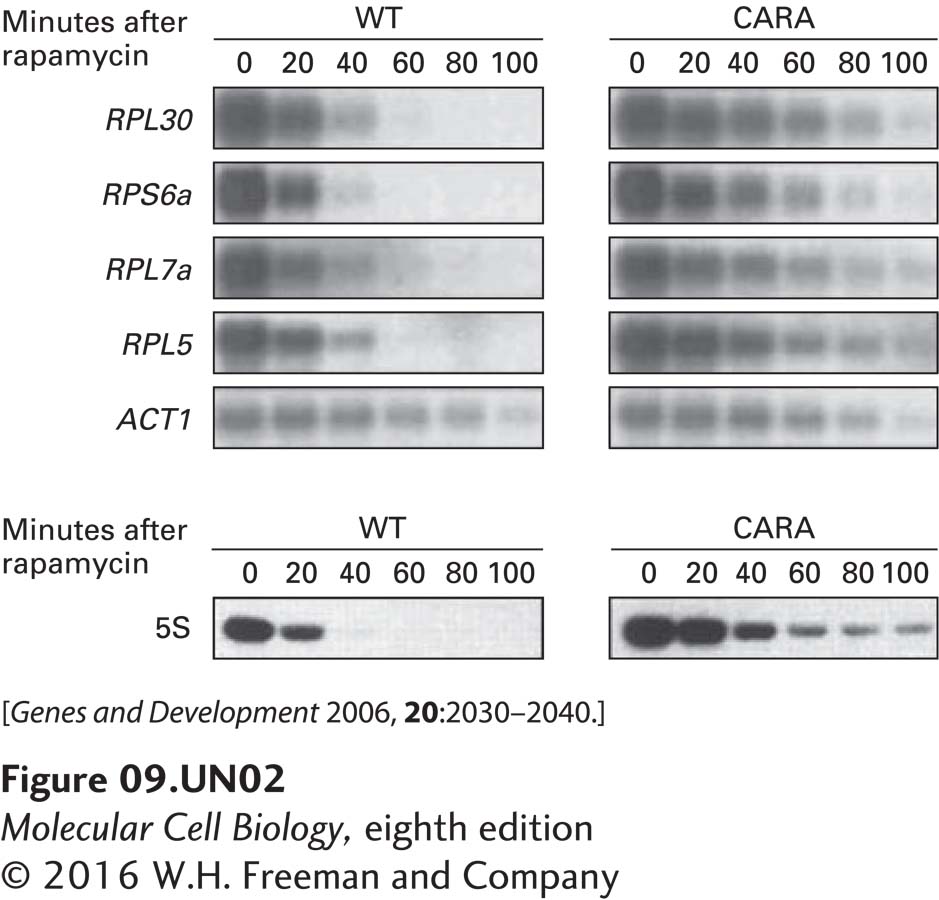
b. The concentrations of four mRNAs encoding ribosomal proteins, RPL30, RPS6a, RPL7a, and RPL5, and of the mRNA for actin (ACT1), a protein present in the cytoskeleton, were assessed in wild-type and CARA cells by Northern blotting at various times after addition of rapamycin to rapidly growing cells (upper autoradiograms). 5S rRNA transcription was assayed by pulse-labeling rapidly growing WT and CARA cells with 3H uracil (for 20 minutes) at various times after addition of rapamycin to the media. Total cellular RNA was isolated and subjected to gel electrophoresis and autoradiography. The lower autoradiogram shows the region of the gel containing 5S rRNA. Based on these data, what can be concluded about the influence of Pol I transcription on the transcription of ribosomal protein genes by Pol II and of 5S rRNA genes by Pol III?
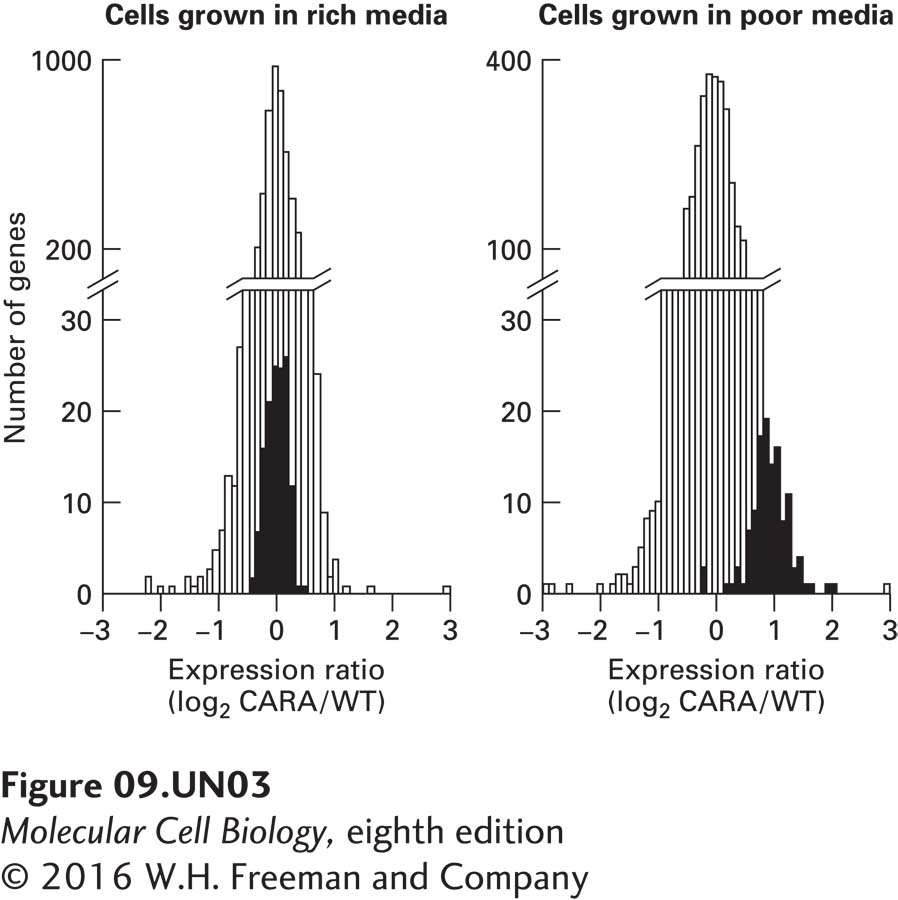
c. To determine whether the difference in the behavior of wild-type and CARA cells could be observed under normal physiological conditions (i.e., without drug treatment), cells were subjected to a shift from nutrient-rich media to nutrient-poor media. Under these conditions, in wild-type cells, the TOR protein kinase becomes inactive. Consequently, shifting cells from nutrient-rich media to nutrient-poor media should result in a normal physiological response that is equivalent to treating cells with rapamycin, which inhibits TOR. To determine how the CARA fusion protein affected the response to this media shift, RNA was extracted from wild-type and CARA cells and used to probe microarrays containing all yeast open reading frames. The extent of RNA hybridization with the arrays was quantified and is expressed in the graphs as log2 of the ratio of CARA-cell RNA concentration to wild-type-cell RNA concentration for each open reading frame. A value of zero indicates that the two strains of yeast exhibit the same level of expression for those specific RNAs. A value of 1 indicates that the CARA cells contain twice as much of that particular RNA as do wild- type cells. The graphs below show the number of open reading frames (y axis) that have values for log2 of each ratio, indicated by the x axis. The results of hybridization to open reading frames encoding mRNAs for ribosomal proteins are shown by black bars, those for all other mRNAs by white bars. The graph on the left gives results for cells grown in nutrient-rich medium, the graph on the right for cells shifted to nutrient-poor medium for 90 minutes. What do these data suggest about the regulation of ribosomal-protein gene transcription by Pol II?
Activity results are being submitted...