 Overall Progress: 0%
Overall Progress: 0%

Chapter 1. Preimplantation Genetic Diagnosis
1.1 Introduction
Preimplantation Genetic Diagnosis
DNA Mutations and Human Disease Case Study
Popup content goes in this box
New box content

Emily Rodgers, having recently finished her PhD in molecular biology, was in the process of interviewing for a laboratory position at IVFTech. IVFTech has pioneered efforts in the field of in vitro fertilization for the past two decades, not only to improve methodologies to raise the success rates of infertile couples, but also to detect genetic mutations that may be present in the preimplantation embryo. She was interviewed by Evan Tucker, who obtained a PhD in genetics and is a genetic counselor by training.
"Good to meet you, Dr Rodgers," he began, "I’m Dr Tucker, but please call me Evan." As they walked into his office, he started talking about the company.
"As you know, about 15% of married couples have issues with reproductive infertility. We use state-of-the-art technologies in the reproductive field so that we can help those couples have children and become parents. At IVFTech, we have a broad array of more than 50 scientists and doctors to achieve this goal including endocrinologists, reproductive biologists, genetic counselors, geneticists, and molecular biologists, to name a few. We work hand-in-hand with regional hospitals so that once a pregnancy is established, the mother and baby are monitored through delivery and beyond."
Emily interjected statistics on how IVFTech has led to a higher success rate amongst infertile couples, and then added, "I also know that your Preimplantation Genetic Diagnosis division has helped to screen for DNA mutations that would give rise to certain human diseases. Early embryos that do not have those mutations then could be used to establish successful pregnancies."
Evan responded, "Exactly! I’m glad you brought that up. The position that I have in mind for you is in the Preimplantation Genetic Diagnosis division, as a section leader in charge of DNA analysis. Let me show you one of our latest cases and we can see how you would do on the job." Emily enthusiastically agreed.
1.2 Allele Inheritance
Allele Inheritance
Evan began, "Many couples come to us for preimplantation genetic diagnosis because they have a family history of a genetic disease, or because they have been identified by genetic testing as carriers of a genetic disease. The first thing we do for these couples is to generate a family history so that we can confirm that their child is at risk. To do this, we have to understand the nature of the genetic mutation, whether it is dominant or recessive."
1.
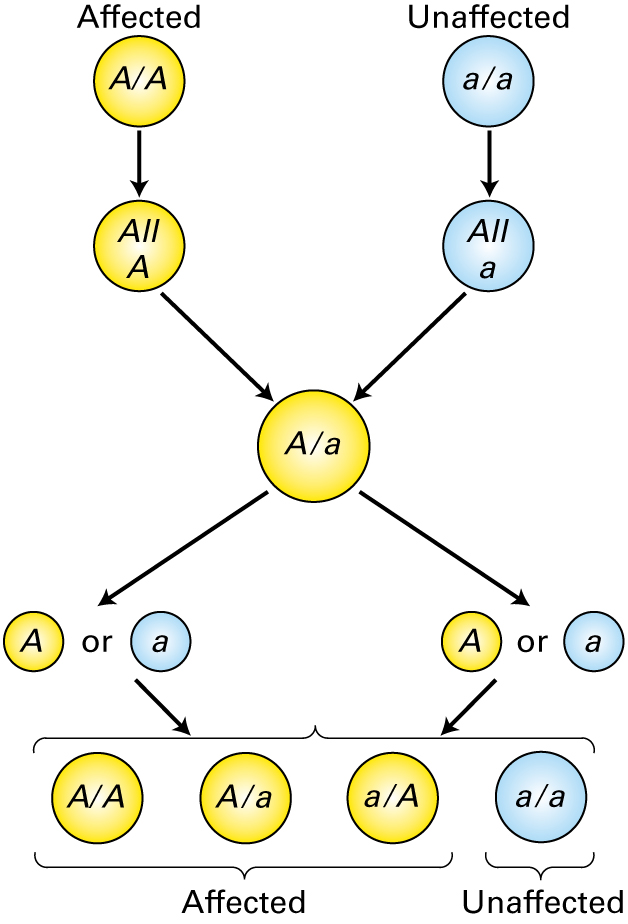
The parental genotypes are shown at the top of this figure. Which statement is true concerning the figure above?
| A. |
| B. |
| C. |
| D. |
| E. |
2.
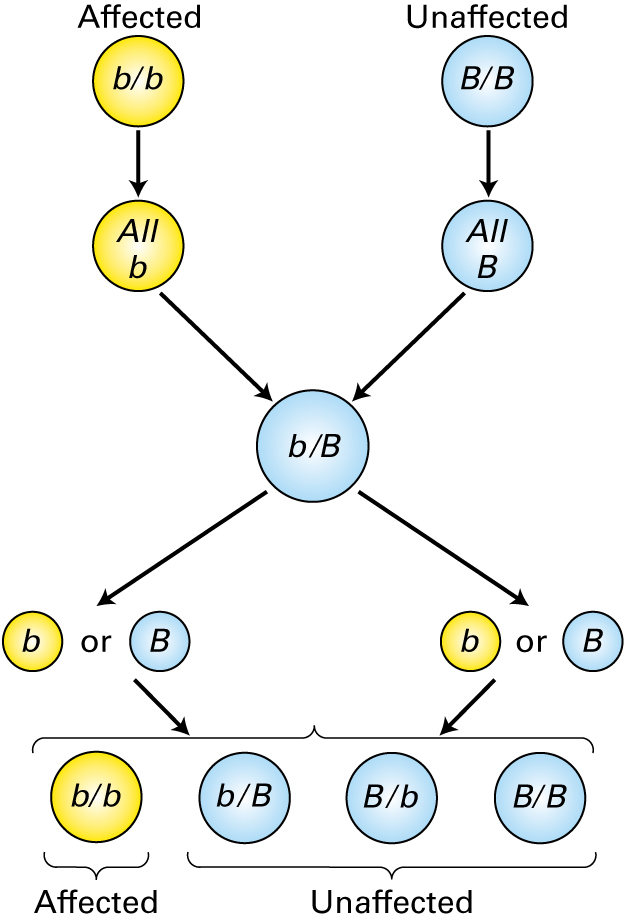
The parental genotypes are shown at the top of this figure. Which statement is true concerning the figure above?
| A. |
| B. |
| C. |
| D. |
| E. |
3.
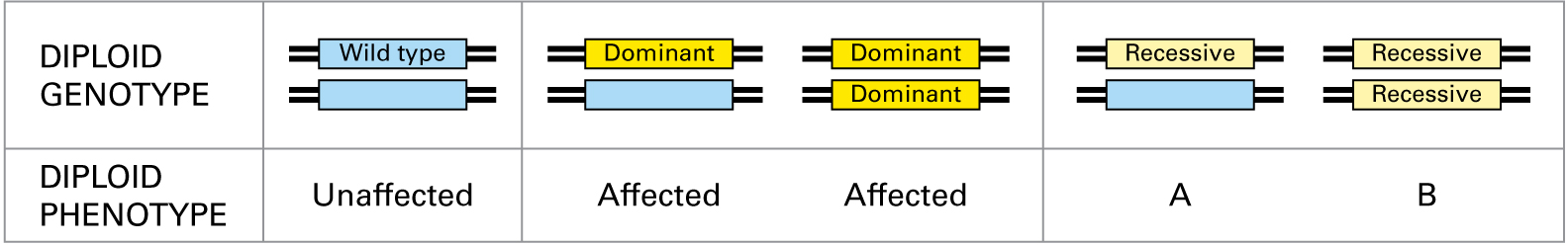
Based on diploid genotypes, the diploid phenotype of situation A would be while the diploid phenotype of situation B would be .
1.3 Family History
Family History
Evan next introduced Family 40513. "The mother," he pointed to the circle labeled (A) on the pedigree below, "had genetic testing before the birth of their first child, and was found to carry a mutation in the CFTR gene. Her husband (B) was also tested, and no mutation was found. Neither of their children has cystic fibrosis, though through testing we learned that the son (F) carries the same mutation as the mother. They came to us because the father’s nephew (G) was recently found to be suffering from cystic fibrosis, but the boy’s mother (C) didn’t appear to be a carrier, based on her prenatal genetic testing. It seems as though there may be a rare or novel CFTR mutation on that side of the family. The couple (A and B) wants to have another child, but they would like to find out if there is a chance their next child will have cystic fibrosis."
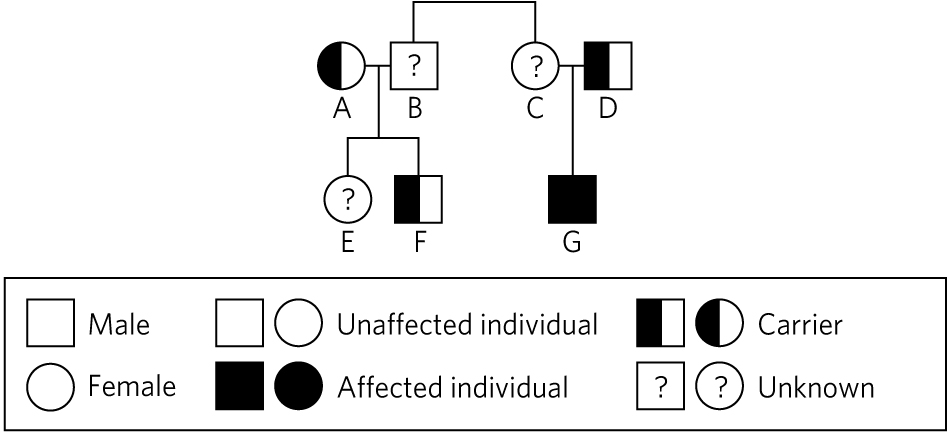
Occurrence of cystic fibrosis in Family 40513.
4.
If the father (B) is NOT a carrier of a CFTR mutation, what is the chance that their next child will be a carrier?
| A. |
| B. |
| C. |
| D. |
| E. |
5.
If the father (B) carries a mutation in CFTR, what is the chance that their next child will also be a carrier?
| A. |
| B. |
| C. |
| D. |
| E. |
6.
If the father (B) carries a mutation in CFTR, what is the chance that their next child will be affected by cystic fibrosis?
| A. |
| B. |
| C. |
| D. |
| E. |
"Very good, Emily," Evan congratulated. "You certainly have the genetics background for this type of work."
1.4 Functional Analysis
Functional Analysis
Evan explained, "Initially, the aunt (C) was screened using a standard cystic fibrosis mutation panel, but no mutation was identified. Since cystic fibrosis was diagnosed in her son (G), we ran an extended CF panel on him and both his parents."

Occurrence of cystic fibrosis in Family 40513.
"Now," Evan folded his hands on his desk. "What do you know about the mutations that cause cystic fibrosis?"
Emily thought for a moment, then answered, "The most common mutation is a deletion of a single phenylalanine amino acid at position 508. I'm afraid I don’t know any others."
"Yes, that mutation is responsible for 70% of cystic fibrosis cases, but there are many other known mutations. Thankfully, a lot of the information we need is available in online databases," Evan reassured her.
Learn about the most common mutations associated with the CFTR gene at http://www.cftr2.org/.
"The IVFTech standard CF panel screens for the seven most common mutations associated with cystic fibrosis," Evan explained, showing Emily the table below.
| Mutation | Exon | Prevalence in CF patients | Cellular effect |
|---|---|---|---|
| F508del | 11 | 70%, highest in Northern European populations | No protein |
| G542X | 12 | 5%, highest in Ashkenazi Jewish populations | No protein |
| G551D | 12 | 4% | Defective gating |
| W1282X | 11 | 4%, highest in Ashkenazi Jewish populations | No protein |
| N1303K | 11 | 2.5% | No protein |
| R553X | 11 | 2.5% | No protein |
| R117H | 11 | 1% | Defective conductance |
7.
"We have designed PCR primers to detect whether those mutations are found in someone’s genomic DNA. Therefore, we have different PCR primer sets, each of which bind within specific exons of the CFTR gene. The mutations shown correspond to the specific alteration at the protein level. For example, F508del refers to the deleted phenylalanine that you mentioned. G551D is a change at position 551 from glycine to aspartic acid. An X refers to a premature stop codon. Different mutations have different effects on the stability and function of the CFTR protein."
Read about the classes of CFTR mutations at http://www.hopkinscf.org/what-is-cf/basic-science/cftr/mutations/.
The CFTR gene encodes for a membrane protein that:
| A. |
| B. |
| C. |
| D. |
| E. |
| F. |
8.
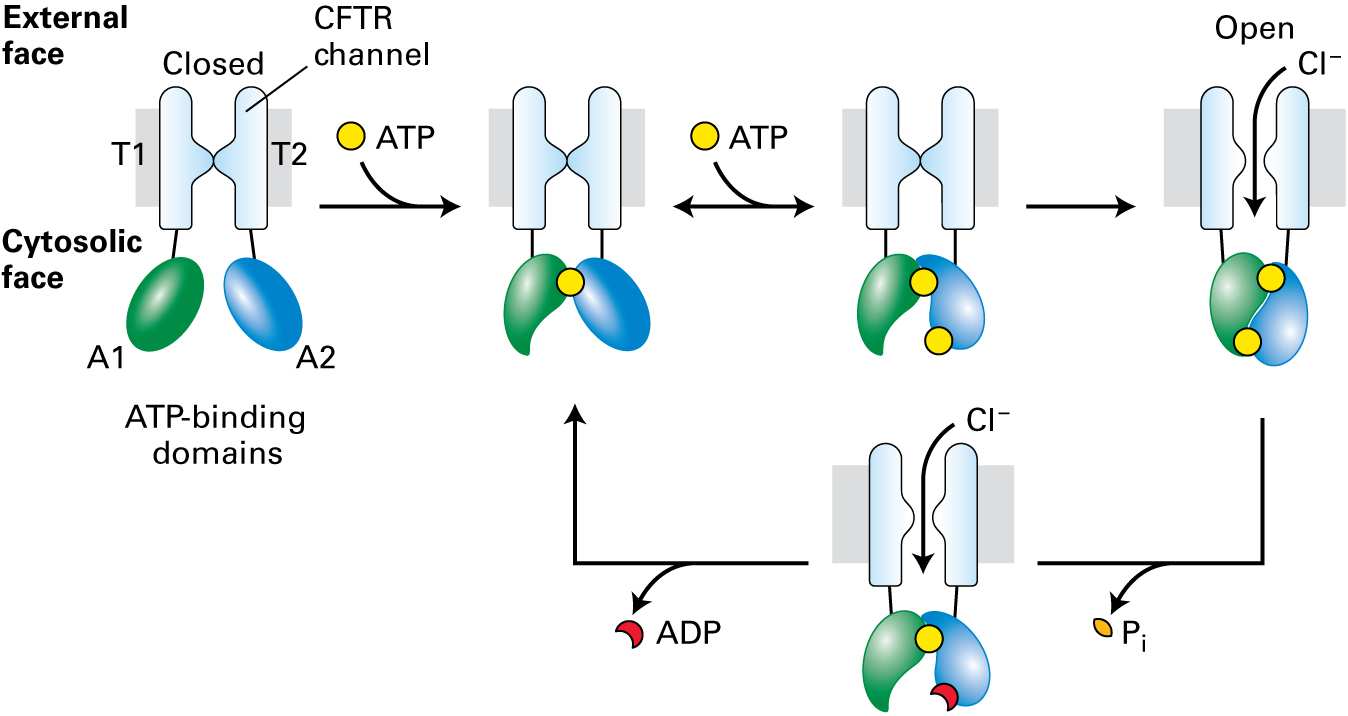
CFTR is a chloride channel expressed in the apical plasma membranes of epithelial cells in the lungs, sweat glands, pancreas, and other tissues. Opening of the channel, composed of two transmembrane domains (T1 and T2), is activated by phosphorylation of the regulatory domain (not shown) and ATP-binding of the ATP-binding domains (A1 and A2).
Cystic fibrosis occurs when the CFTR channel fails to move chloride ions across the epithelial cell membrane, causing an imbalance of ions and multiple downstream effects. Which types of mutations in the CFTR gene would NOT result in cystic fibrosis?
| A. |
| B. |
| C. |
| D. |
| E. |
9.
Which mutation class would be least severe in terms of phenotype?
| A. |
| B. |
| C. |
| D. |
| E. |
The most common mutation, the F508del mutation, causes the protein to fold improperly and be degraded before it reaches the plasma membrane.
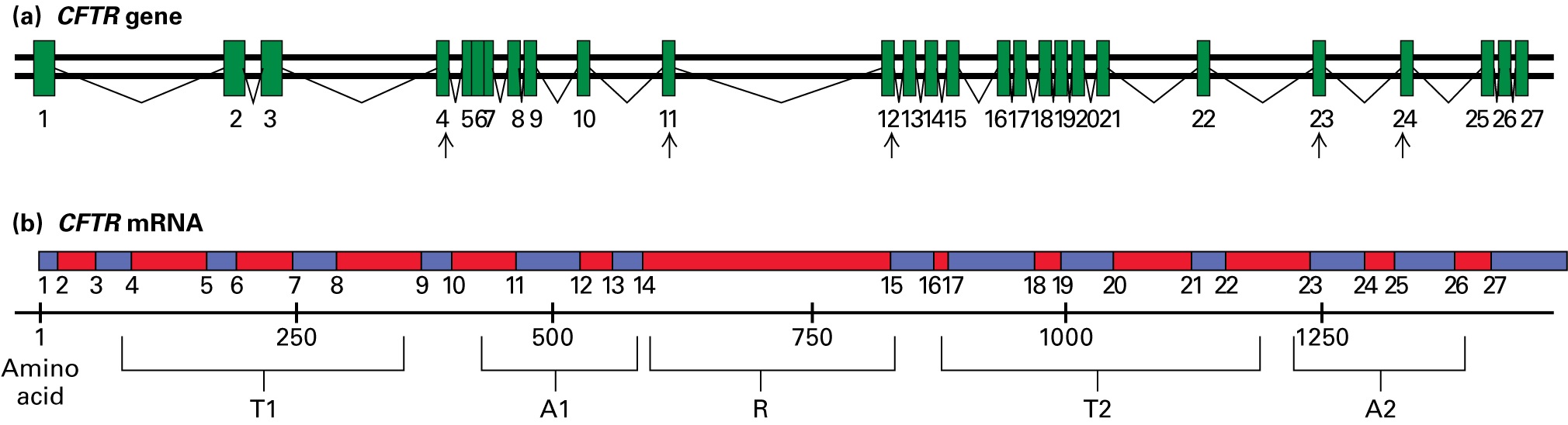
Evan gave Emily more details about the CFTR gene and its protein product. "The CFTR gene is 230,000 base pairs long, consists of 27 exons, and creates a protein that is 1480 amino acids long. This figure depicts the CFTR gene and its mRNA, showing which amino acids encode the protein domains: two transmembrane segments (T1 and T2), two ATP-binding domains (A1 and A2), and the regulatory domain (R).
"Arrows point to areas where we have exon-specific primers that we use to detect the most common mutations: Exon 4–specific primers for the R117H mutation; exon 11–specific primers for the F508del mutation; exon 12–specific primers for the G542X, G551D, and G553X mutations; exon 23–specific primers for the W1282X mutation; and exon 24–specific primers for the N1303K mutation. We also have other primer sets developed so that we can amplify any of the exons from genomic DNA."
"The nephew (G) in family 40513 was found to have the F508del mutation in one allele, which his father (D) also carries. However, neither our standard CF panel, nor the extended CF panel, was able to identify a mutation in the allele he inherited from his mother. Because we don’t know what CFTR mutation this family carries, we can’t easily determine whether the father (B) is a carrier."

Occurrence of cystic fibrosis in Family 40513.
Emily spoke up, "You must be able to identify the mutation via sequencing, right?"
Evan nodded, "Yes, we could sequence the entire CFTR gene, but that would take some time and even then, we might not find the mutation if it’s in an upstream or downstream regulatory region. Our team began working to identify the mutation right away, and I’ll show you what they found a little later. This family wanted to have another child soon, so they opted to take advantage of our IVF and preimplantation diagnosis services."
"Oh!" Emily exclaimed, "Because cystic fibrosis is recessive, all you have to do is find an embryo that doesn’t have the mother’s mutation!"
"That’s right," Evan said. "If the embryo doesn’t have the maternal mutation, then it most likely inherited the unaffected allele from its mother, and it will either be a carrier, like the father, or completely unaffected."
1.5 Diagnosis
Diagnosis
So far, Evan was impressed with Emily’s analysis. He continued telling her about Family 40513. "The parents came in for IVF treatment, and we did an assessment of the embryos to find out the genotypes of the CFTR alleles for each.
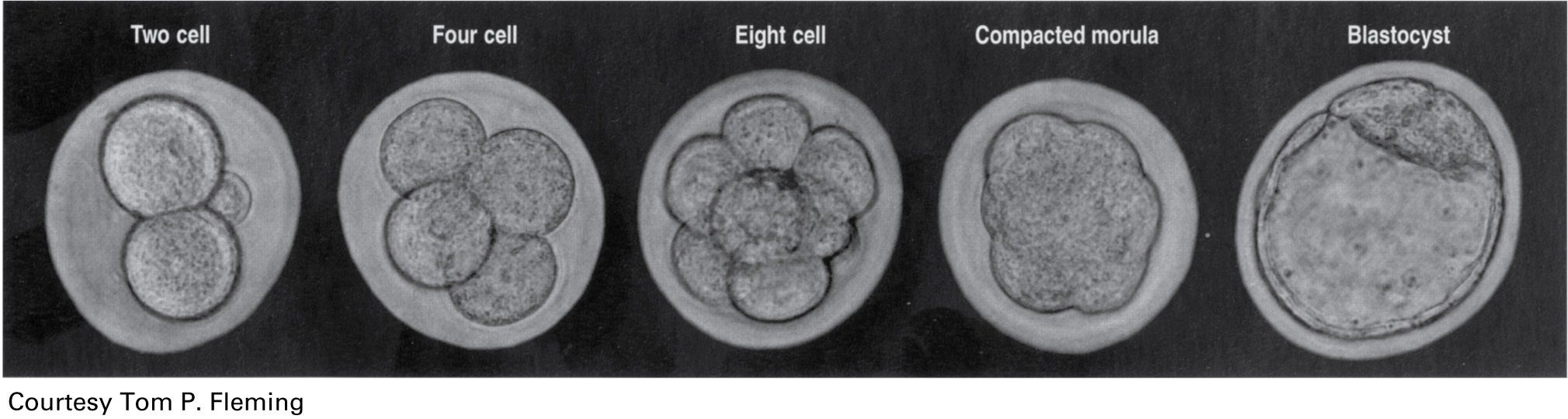
"From the in vitro fertilization procedure, the early embryo is cultured through successive stages shown here. At the eight-cell stage (middle figure in panel below) each cell is referred to as a blastomere. A single blastomere (cell) can be gently removed for testing, leaving the other seven cells behind, and the embryo is then cultured to the blastocyst stage (last figure in the panel below). At this point, the blastocyst is frozen until our tests are completed. Those that do not have the mutant allele can be thawed and used to establish a successful pregnancy. The normal development of the human embryo from the single cell stage through the blastocyst stage takes approximately four and a half days.
10.
"We were able to obtain a single blastomere from eight different early embryos. Since we are starting from a single cell in each case and are limited with the amount of DNA, what is the best approach to analyze the genomic DNA?"
| A. |
| B. |
| C. |
| D. |
| E. |
11.
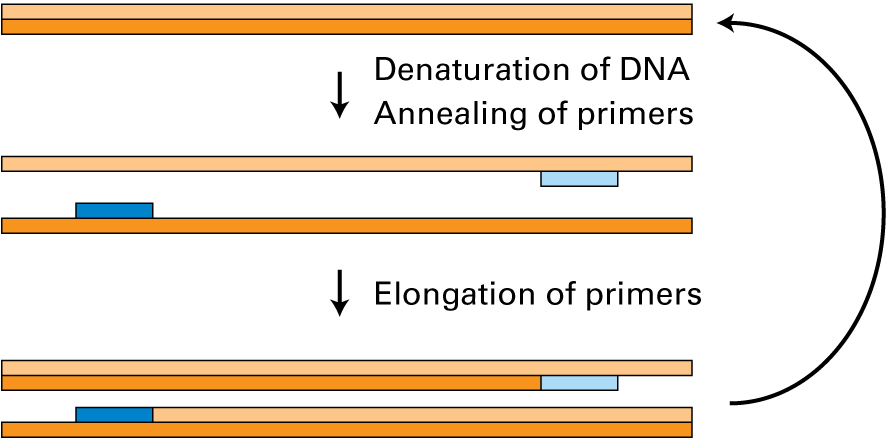
There are generally three distinct temperatures that are used in PCR. Which of the following temperatures are correctly paired with the corresponding PCR events?
| A. |
| B. |
| C. |
| D. |
| E. |
12.
Evan continued, "We ran our standard CF screen for the seven most common CF mutations on each embryo. Using the gene-specific primers to amplify each embryo’s genomic DNA at exons 4, 11, 12, 23, and 24, we got these PCR results.”

What can you conclude about the genotypes of the embryos with regard to the CFTR gene? Is this expected from the nature of the mutations tested? Why or why not?
13.
What would be the best method to detect specific base changes in DNA?
| A. |
| B. |
| C. |
| D. |
| E. |
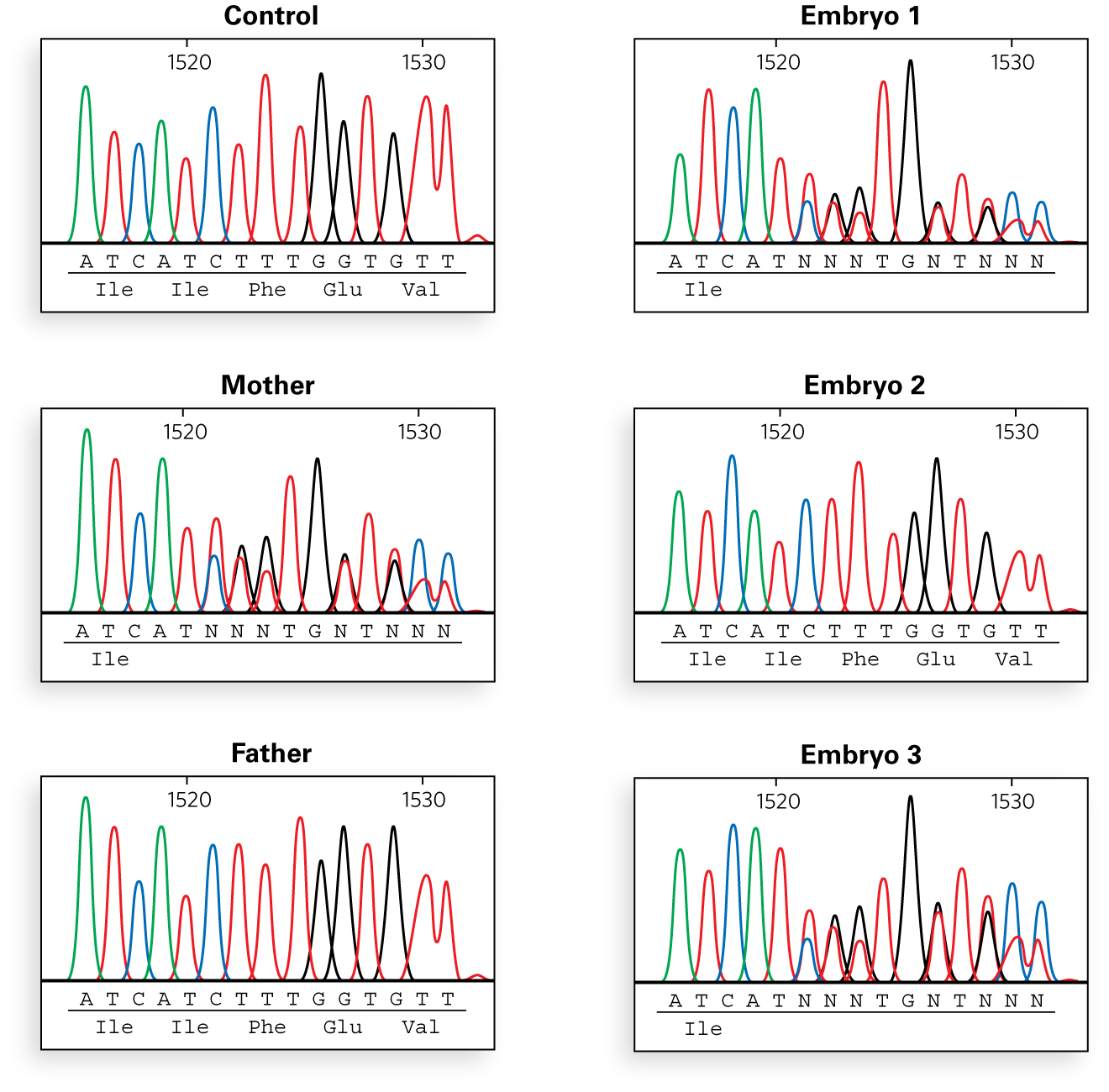
Evan explained, "We sequenced the PCR products from the mother, father, and eight embryos to look for small changes in the DNA sequence. No differences were found in the sequences for exons 4, 12, 23, and 24, compared with the control. This is what we found in exon 11.
14.
What is the nature of the mutation on the mother’s side of the family?
| A. |
| B. |
| C. |
| D. |
| E. |
Correct.
Incorrect.
Try Again.
"So, you can see from the sequencing results,"Evan said, "that the mother’s mutation is the deletion of phenylalanine 508, and that embryos 1 and 3 carry that mutation. Embryo 2 does not have that deletion and would likely be either a carrier of the paternal mutation or completely unaffected. However, we were able to identify the mutation on the father’s side by the time the sequencing results came back, before implantation. Let me show you how.”
1.6 Mutation Identification
Mutation Identification
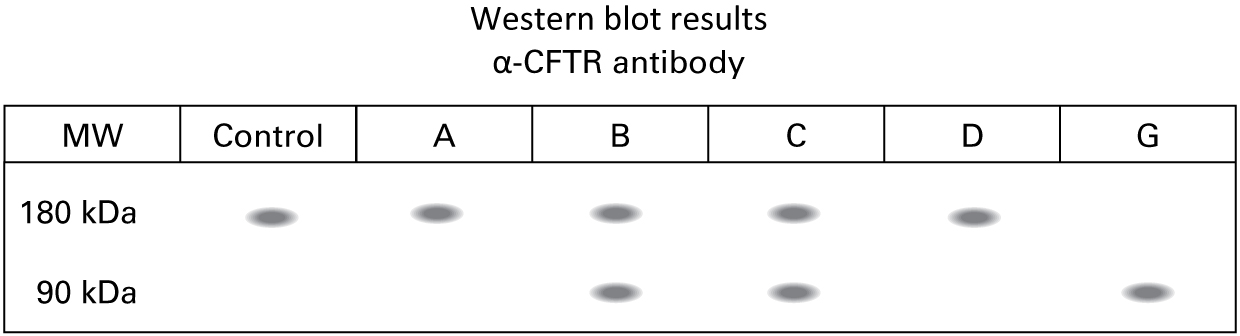
Evan flipped to a new page in the case file. "We knew the nephew (G) inherited the F508del from his father (D) and likely an unknown mutation from his mother (C). We were able to obtain lung biopsies from the whole family. We isolated protein from those tissues and ran a Western blot using an N-terminus specific antibody against CFTR. Here are the results."

15.
What can you conclude about the mutation on the father’s side of the family? Which family member(s) carry this mutation?
16.
"We also ran PCR on all the exons, and found the following results when using a primer set designed to amplify exon 15."
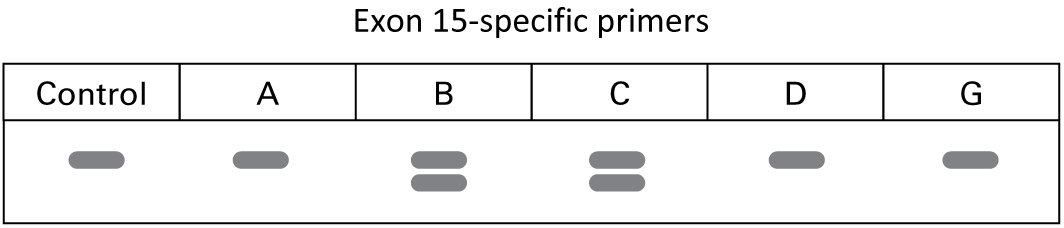
What is the most likely type of mutation along the father’s side of the family?
| A. |
| B. |
| C. |
| D. |
| E. |
17.
Evan cut to the chase. "Sequence analysis of the smaller PCR fragment reveals that there was a large deletion within exon 15, causing a frameshift that resulted in a premature stop codon and truncated protein. After assessing DNA sequencing results, the following genotypes were assigned to the embryos."
| Embryo 1: F508del/ exon 15del | Embryo 2: exon 15del/+ | Embryo 3: F508del/+ |
| Embryo 4: +/+ | Embryo 5: +/+ | Embryo 6: F508del/ exon 15del |
| Embryo 7: F508del/ exon 15del | Embryo 8: +/+ |
Which embryo would be considered a carrier?
| A. |
| B. |
| C. |
| D. |
| E. |
Evan concluded, "Embryos #4 (male) and #5 (female) were successfully implanted, and the couple are expecting two new additions to the family in about eight months."
"Gee, this sounds like a really interesting job!" Emily exclaimed.
Evan smiled and said, "I’m glad you think so. I think you’ll be a good fit with the rest of our team. Do you have any questions about the job?"
Emily asked, "I read on the IVFTech website that you have a research and development division. Can you tell me about your research goals?"
1.7 Gene Therapy
Gene Therapy
"Absolutely," Evan replied. "Although our number one goal is to prevent any serious genetic diseases, we realize that many people are living with these diseases already. Our research and development division is working on methods to treat or cure those diseases using gene therapy. In order to perform gene therapy in humans, first we need to perform experiments in human cell lines and perhaps in animal models of human disease to demonstrate proof in principle. We have state-of-the-art transgenic and knockout facilities to be able to make mouse models of human diseases."
18.
Based on the nature of cystic fibrosis, would you recommend a transgenic overexpressing mouse model or a gene knockout mouse model to help characterize the gene? (Hint: See Chapter 6, p. 259.)
19.
We have mouse knockout models for cystic fibrosis, but in some cases the phenotype manifested in multiple tissues, making it difficult to discern the contribution of the mutant allele to lung physiology.
What method could you use to generate a more refined model that only addresses the phenotypic effects in the lung? That is, how would you perform a gene deletion only in the lung and not globally in every cell?
| A. |
| B. |
| C. |
| D. |
| E. |
Correct.
The Cre-lox system is a specific recombination system isolated from bacteriophage P1 that is extremely useful in the genetic engineering field to perform tissue-specific deletion experiments. Cre is a recombinase protein that binds to its recognition site, the 34-bp loxP sequence. When two loxP sites are placed around a gene of interest, Cre will bind to the loxP sites and delete the intervening sequence. A similar system, the FLP-FRT system, was isolated from the yeast 2-micron plasmid and is also used for site-specific recombination in mammalian cells.
Incorrect.
The Cre-lox system is a specific recombination system isolated from bacteriophage P1 that is extremely useful in the genetic engineering field to perform tissue-specific deletion experiments. Cre is a recombinase protein that binds to its recognition site, the 34-bp loxP sequence. When two loxP sites are placed around a gene of interest, Cre will bind to the loxP sites and delete the intervening sequence. A similar system, the FLP-FRT system, was isolated from the yeast 2-micron plasmid and is also used for site-specific recombination in mammalian cells.
Try Again.
Evan went on to explain their approach to gene therapy. "In our group, we have developed mouse models for several human diseases not only to understand the underlying pathologies, but to use them in gene therapy–based approaches. The two basic strategies are: (1) direct delivery, if there is access to the cells that need correction, and (2) cell-based delivery. With respect to cystic fibrosis, we have generated mouse models for the seven most common CFTR human mutations, and we use them to test our gene therapy-based treatment strategies.
"The direct delivery method is preferred for delivering and integrating a functional CFTR gene into the patient’s lung cells, since the respiratory epithelial cells are readily accessible. The gene is in the form of a cDNA, much smaller than the genomic DNA, which is made from the mRNA, and does not contain intronic sequences. Since viruses are traditionally used to package DNA and have limitations in packaging size, the cDNA is commonly used. The cDNA is packaged into an expression vector, which is then encapsulated by a nonviral synthetic liposome. These liposomes are then aerosolized to be able to reach the target tissue. Cells that take up the liposomes are then able to express protein from the introduced cDNA."
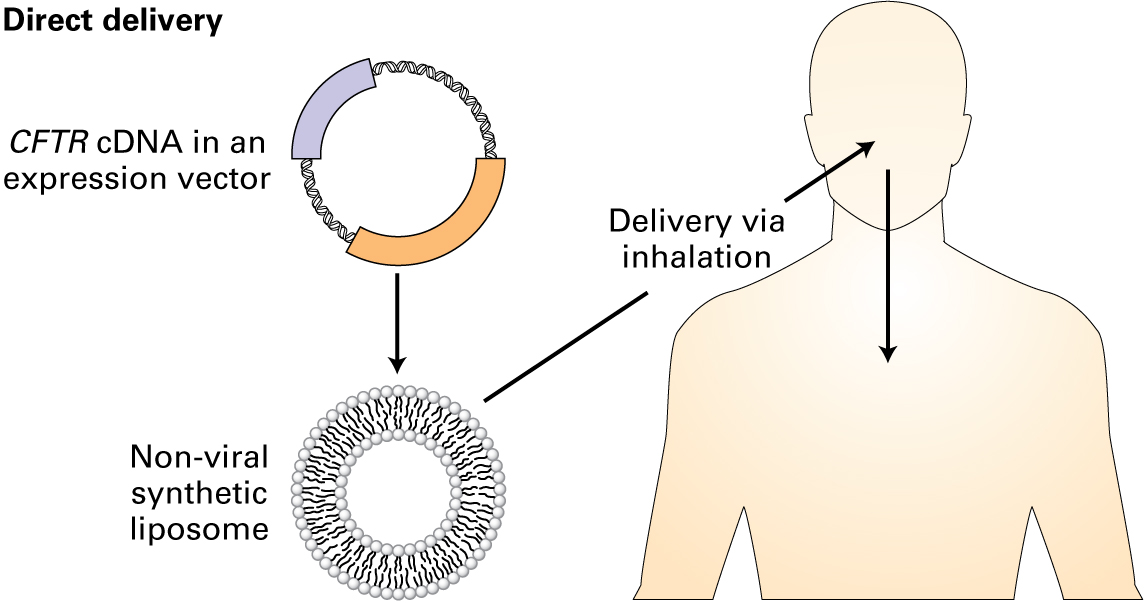
"That sounds like a great advancement for people with cystic fibrosis," Emily said. "And you could potentially use that delivery system to deliver a functional gene in other loss-of-function diseases."
"That's right," Evan said. "For diseases that have a dominant mutation, we are using a cell-based delivery system to initially isolate cells, then perform gene editing to correct the DNA mutations, and administer them back into the patient."
20.
What gene editing technique would be an efficient way to modify an existing malicious gene in somatic cells?
| A. |
| B. |
| C. |
| D. |
| E. |
"Thanks for coming in, Emily. I look forward to working with you."

Your grade of 0% has been submitted.