 Overall Progress: 0%
Overall Progress: 0%

Chapter 1. To Kill A Cancer Cell
1.1 Introduction
To Kill A Cancer Cell
Signal Transduction Case Study
Popup content goes in this box
New box content

As the lead scientist at the largest pharmaceutical company in North America, PharmUSA, Kaitlin Summers is in charge of developing new drugs to combat cancer. It has been a long year, but her team has developed several new drugs for preclinical testing in cell lines, and even more are in the pipeline. Because she frequently partners with colleagues in academia on projects, she decides to give one of her old lab mates, Rick Stanton, a call. She and Rick got PhDs in the same lab at Cal State.
After spending a few minutes catching up on their personal lives, they start talking science. Rick remarks, “We've generated some data from pancreatic tumor samples that we've been collecting over the past year."
He continues, “We've got nearly 100 samples and have generated cell lines from most of them. Almost 80 percent of these cancer cell lines are secreting an uncharacterized protein we're calling 'protein X.' It's a 90-kDa protein that appears to stimulate cell proliferation and survival. The pancreatic cancer cell lines expressing this protein are from those cancers that have shown metastases, so they are more aggressive than the cell lines that don't express it. If we can figure out the signaling pathway that's responsible for its expression … "
Kaitlin finishes his sentence “ … then we can target that pathway with an inhibitor drug and kill the cancer cells."
Rick agrees to send 10 pancreatic cancer cell lines that express protein X, 2 pancreatic cancer cell lines that do not express protein X, and a cell line derived from normal pancreatic tissue.
After getting off the phone, Kaitlin sets up a conference call with the research heads of four divisions that are looking at different signaling pathways, and they collectively agree to convene the following day to discuss the project.
1.
What question is Kaitlin's research group trying to answer?
The next day, Kaitlin’s research group convenes. Amber Collins leads the receptor tyrosine kinase group. Tom Welch is head of the G protein–coupled receptor group. Dan Tucker is in charge of the TGF-β signaling group. Sarah Wentworth leads the Wnt signaling team.
After the group discusses Rick’s preliminary findings with the pancreatic cancer cell lines, they decide that the best approach is to identify the signaling pathway responsible for the expression of protein X. Once that pathway is determined, the drug development team can test pathway inhibitors to selectively kill the cancer cells. Kaitlin wishes the team good luck before they adjourn.
Each of the labs is set up for the following protocols:
- PCR (polymerase chain reaction): A technique for amplifying a specific DNA segment in a complex mixture by means of multiple cycles of DNA synthesis from short oligonucleotide primers followed by brief heat treatment to separate the complementary strands (see Figure 6-18)
- RT-PCR: Amplification of a specific mRNA in a complex mixture using a reverse transcriptase step followed by PCR (see Figure 6-19)
- Western blotting: Identification of a specific protein in a complex mixture by means of gel electrophoresis and specific antibodies (see Figure 3-41)
- Immunoprecipitation: A technique that uses antibodies to separate a target molecule of interest from other molecules in a complex mixture in solution by cross-linking the target molecule into a large aggregate, resulting in the formation of an insoluble solid (precipitate) that can be easily separated and analyzed (see Figure 3-41)
- Immunofluorescence microscopy: A technique used to localize a specific protein in a cell using an antibody with a fluorescent tag (see page 144, Figure 4-14)
1.2 Amber's lab: Receptor tyrosine kinase pathways
Amber's lab: Receptor tyrosine kinase pathways
After a few days, Kaitlin stops by Amber's lab to check in. Amber gives the following report:
"Good morning! During our analyses of these cells, we've found identical results in all 10 of the aggressive pancreatic cancer cell lines expressing protein X. If we grow the cells to the point that they form a monolayer that covers the plate, they really start to express protein X more robustly. We've had to keep them at about 50 percent coverage to dissect out contributions from different signaling pathways. The 2 nonaggressive pancreatic cancer cell lines have also shown identical results. Dan, Tom, and Sarah have found the same to be true in their experiments. To simplify the data analysis, it's easier to look at the results from each research group. Let's look at what we've found."

1. PCR
"We did not do PCR, but we did do RT-PCR assays."

2. RT-PCR
"We isolated RNA from the cells, made cDNA using a reverse transcriptase protocol, and then used primers specific for HERs (human epidermal growth factor receptor tyrosine kinases) in an RT-PCR assay. Here are the data."
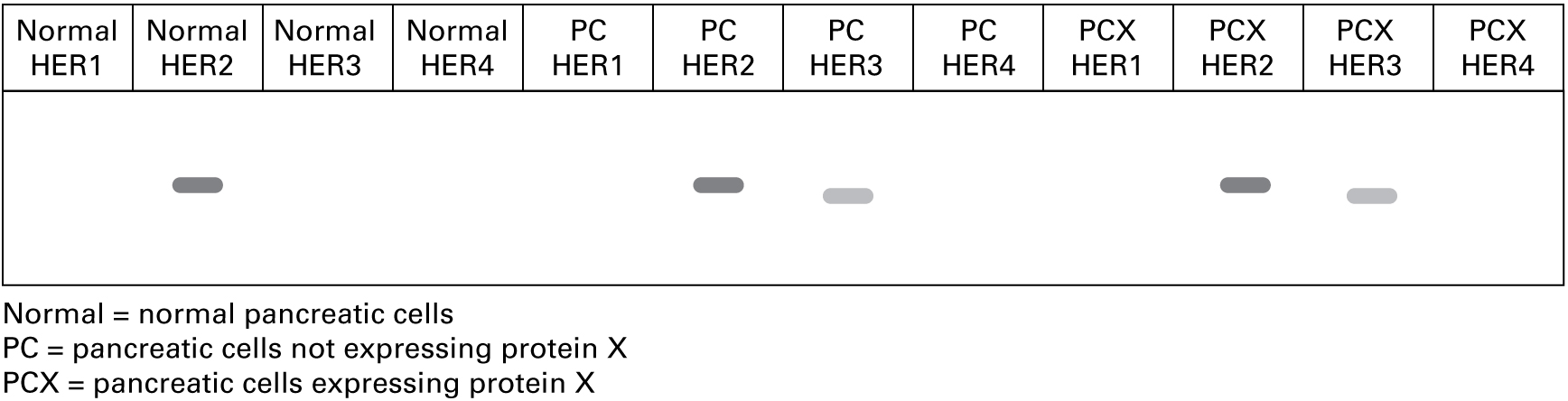

3. Western blot
"We isolated proteins from the cells, ran protein samples out on 8 percent polyacrylamide gels, and transferred the proteins to nitrocellulose membranes for blotting with HER-specific antibodies. Based on the RT-PCR data, which antibody or antibodies do you think we tested?"
2.
| a. α-HER1 antibody | YesNo |
| b. α-HER2 antibody | YesNo |
| c. α-HER3 antibody | YesNo |
| d. α-HER4 antibody | YesNo |
Correct.
"Since the RT-PCR data showed expression of HER2 and HER3, we performed two Western blots, one using the α-HER2 antibody and the other using the α-HER3 antibody. The expected molecular weight of HER2 is approximately 138 kDa, and the expected molecular weight of HER3 is about 135 kDa."
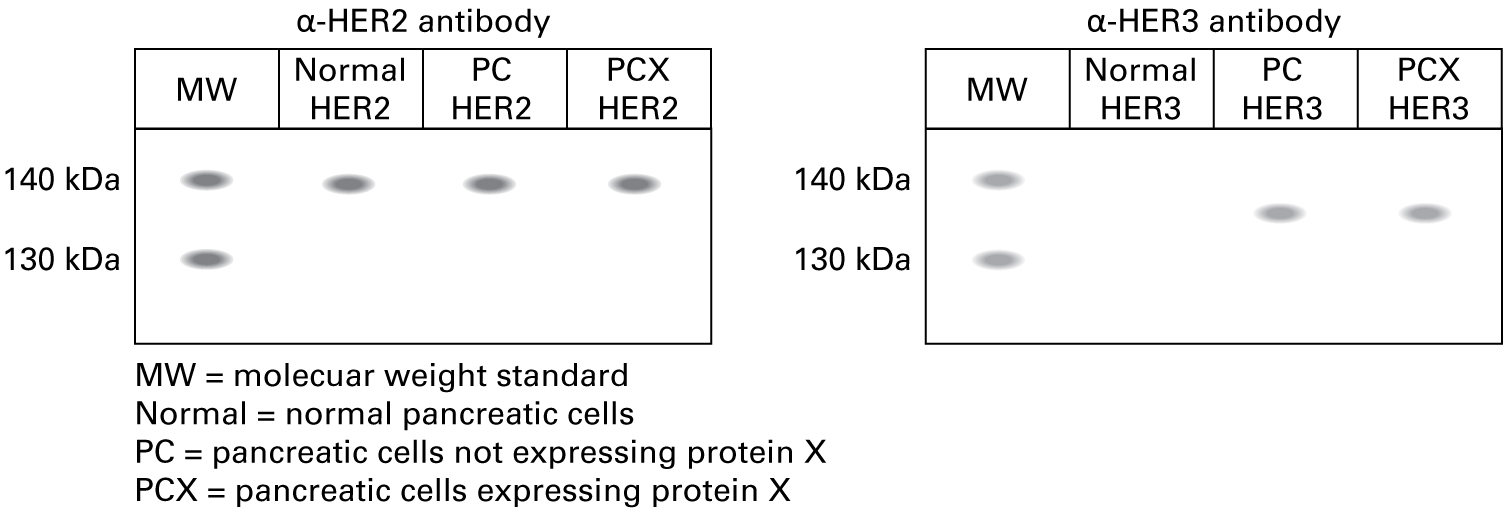
Incorrect.
"Since the RT-PCR data showed expression of HER2 and HER3, we performed two Western blots, one using the α-HER2 antibody and the other using the α-HER3 antibody. The expected molecular weight of HER2 is approximately 138 kDa, and the expected molecular weight of HER3 is about 135 kDa."

3.
There are four HERs (HER1, HER2, HER3, and HER4). Once engaged by a ligand, what are the dimerization possibilities for HER3?
| A. |
| B. |
| C. |
| D. |
| E. |
| F. |
4.
Which two experiments together would reveal whether HER2 and HER3 are forming heterodimers?
| a. First experiment: | |
| b. Second experiment: |

4. Immunoprecipitation
5.
Given our previous results, which two antibodies would be the most useful for looking at heterodimerization?
| A. |
| B. |
| C. |
| D. |
| E. |
| F. |
Correct.
"We isolated proteins from the cells and used either the α-HER2 antibody or the α-HER3 antibody for immunoprecipitation. Each precipitated protein fraction was run on 8 percent polyacrylamide gels, and proteins were transferred to nitrocellulose membranes for blotting with either the α-HER2 antibody or the α-HER3 antibody. Here are the data."
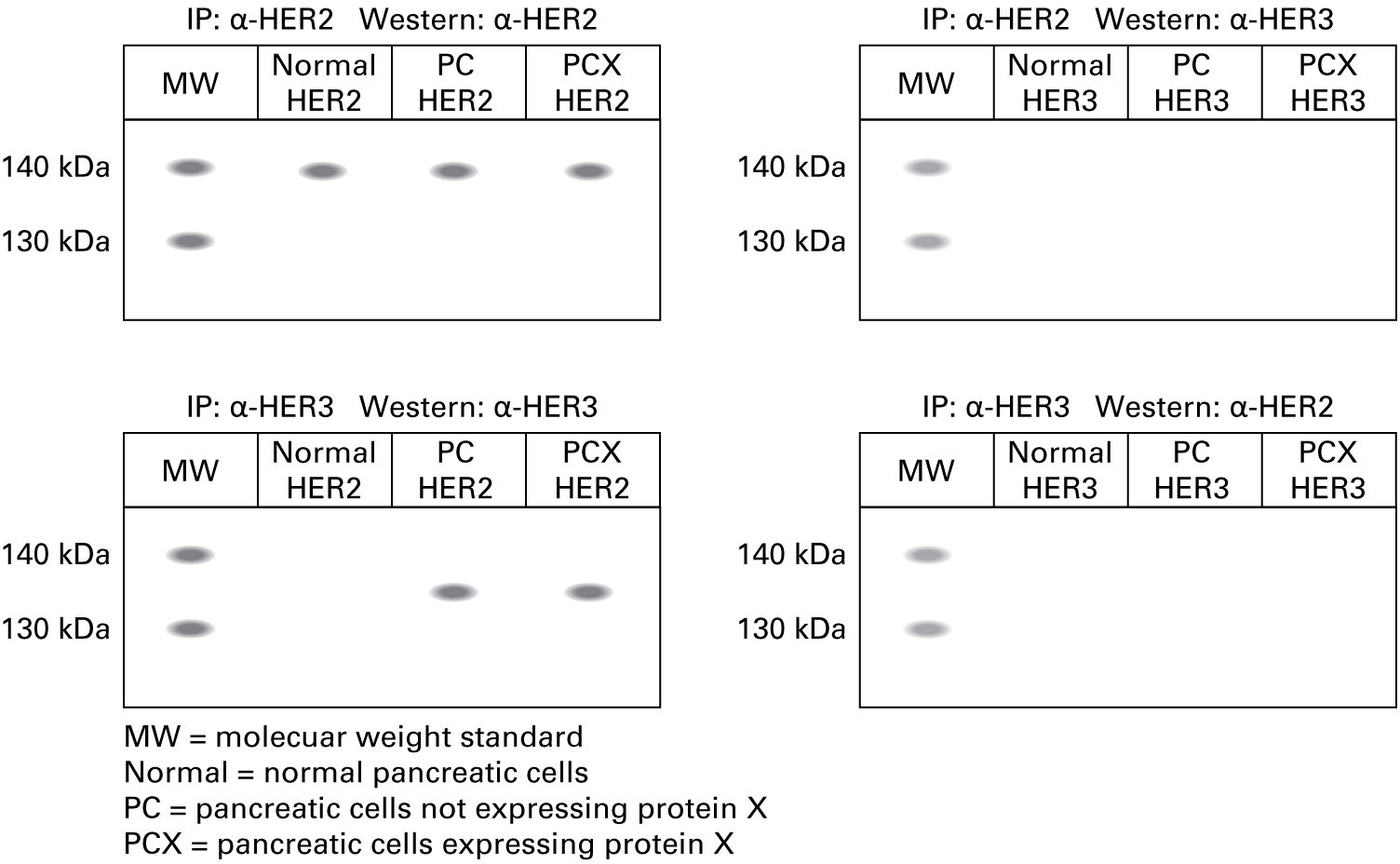
Incorrect.
"We isolated proteins from the cells and used either the α-HER2 antibody or the α-HER3 antibody for immunoprecipitation. Each precipitated protein fraction was run on 8 percent polyacrylamide gels, and proteins were transferred to nitrocellulose membranes for blotting with either the α-HER2 antibody or the α-HER3 antibody. Here are the data."


5. Immunofluorescence microscopy
"We used fluorescein-conjugated antibodies for HER2 and a rhodamine-conjugated antibody for HER3. Here are the data."
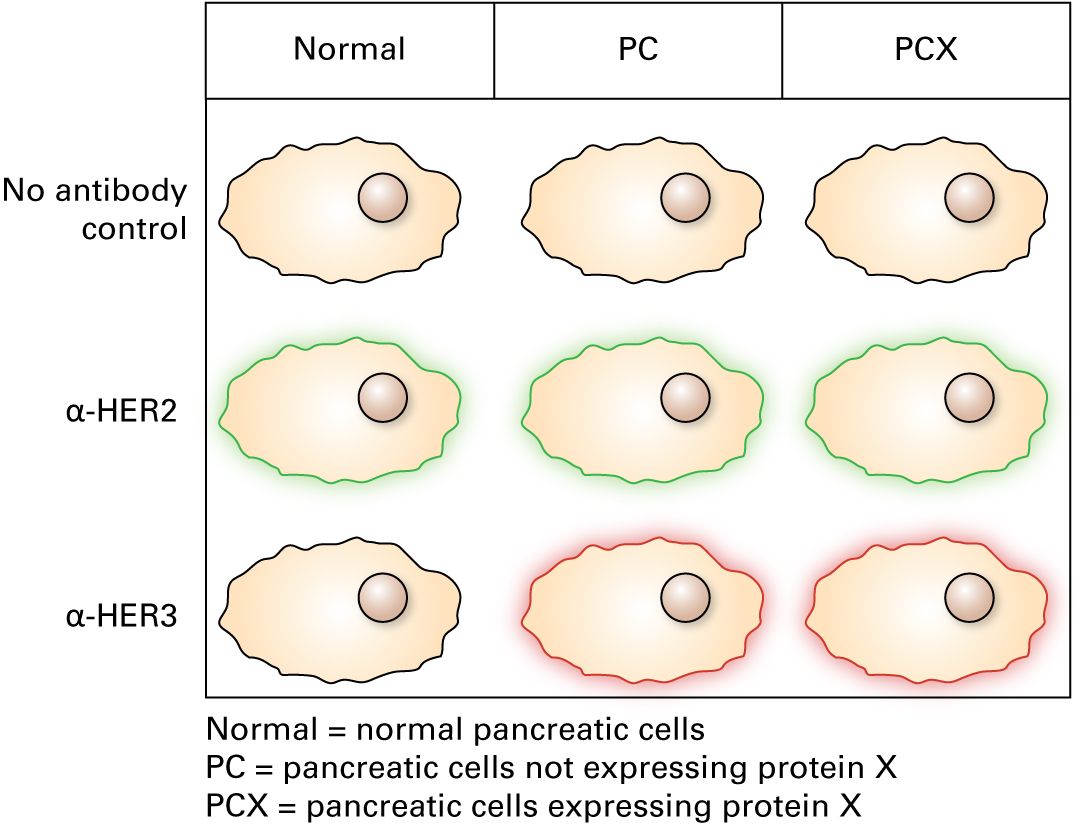
6.
What are the results from the experiments (RT-PCR, Western blotting, co-immunoprecipitation, and immunofluorescence microscopy)?
The RT-PCR data show expression of HER2 mRNA in all cell types and of HER3 mRNA in both cancer cell types.
Western blotting shows the same pattern of expression: HER2 protein in all cell types, HER3 protein only in cancer cell types.
Co-immunoprecipitation does not reveal any interactions between HER2 and HER3 monomers.
Immunofluorescence microscopy shows that each receptor is present on the plasma membranes of the cell types in which it is expressed.
7.
Based on the results to date, the team concludes that HER2 and HER3 are both present on the plasma membranes of both cancer cell types, but do not heterodimerize. To test the ability of a ligand to induce dimerization of these receptors, and subsequent cell signaling, which of the following ligands would you use?
| A. |
| B. |
| C. |
| D. |
| E. |
| F. |
1.3 Tom's lab: G protein-coupled receptor pathways
Tom's lab: G protein-coupled receptor pathways
Kaitlin next stops by Tom's lab. Tom reports:
"We've looked at some of the G protein–coupled receptors in all of the cell types. Adrenergic receptors are expressed in the cells, and it seems that they preferentially have the β-adrenergic receptor and also have cytoplasmic cyclic AMP. Let's think about what may be going on."

1. PCR
"We did not do PCR, but we did do RT-PCR assays."

2. RT-PCR
"We isolated mRNA from the cells, made cDNA using a reverse transcriptase protocol, and then used GPCR-specific primers in an RT-PCR assay. Here are the data."
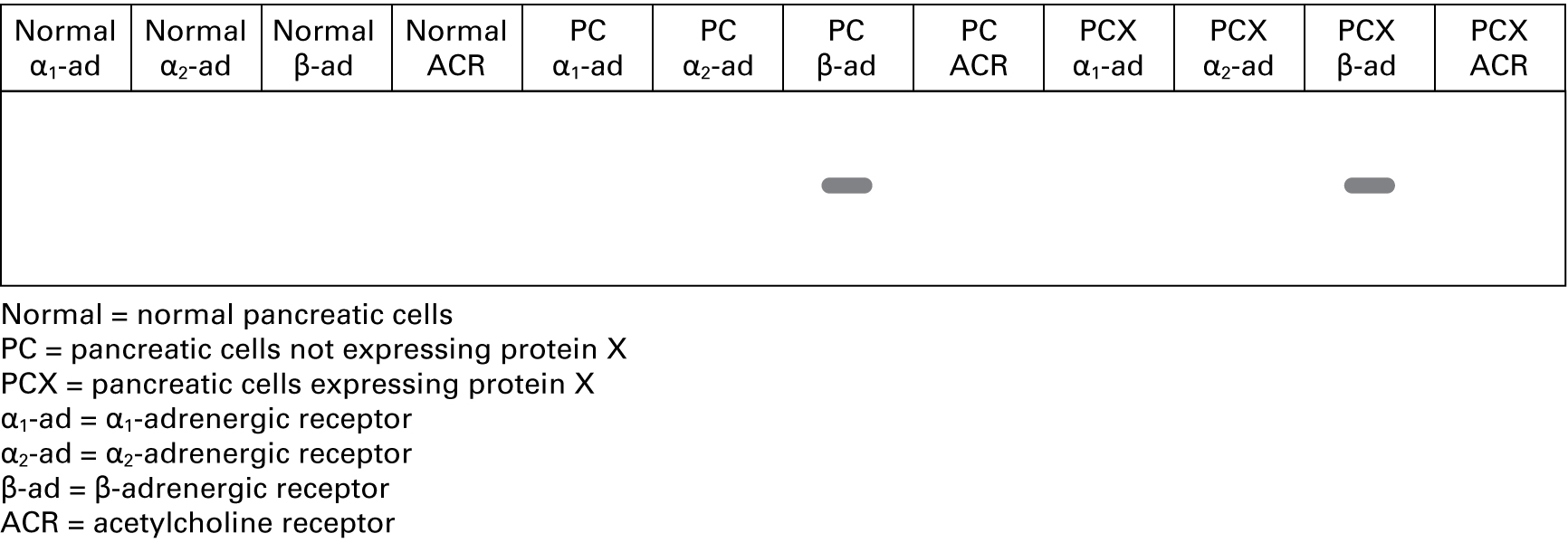

3. Western blot
"We isolated proteins from the cells, ran protein samples out on 8 percent polyacrylamide gels, and transferred the proteins to nitrocellulose membranes for blotting with GPCR-specific antibodies. Based on the RT-PCR data, which antibody or antibodies do you think we tested?"
8.
| a. α-α1-ad antibody | YesNo |
| b. α-α2-ad antibody | YesNo |
| c. α-β-ad antibody | YesNo |
| d. α-ACR antibody | YesNo |
Correct.
"Since the RT-PCR data showed expression of the β-adrenergic receptor, we performed a Western blot using the α-β-ad antibody. The expected molecular weight of the β-adrenergic receptor is approximately 48 kDa."
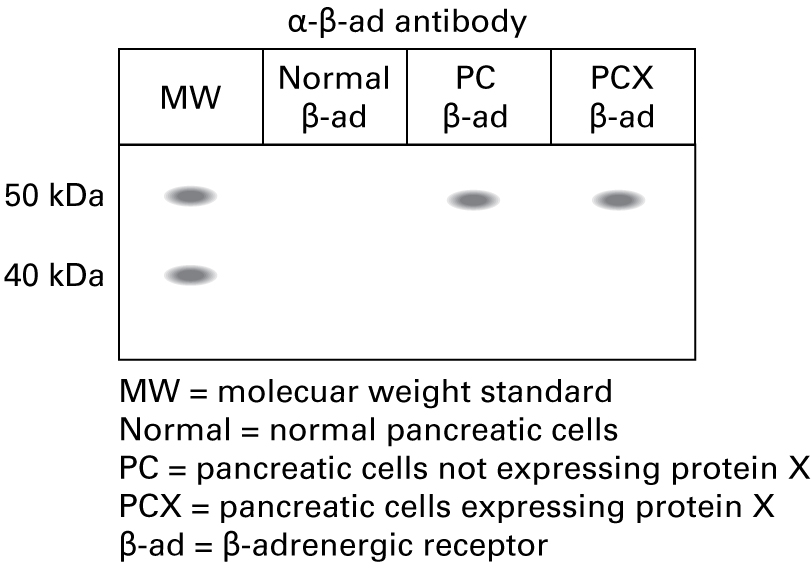
Incorrect.
"Since the RT-PCR data showed expression of the β-adrenergic receptor, we performed a Western blot using the α-β-ad antibody. The expected molecular weight of the β-adrenergic receptor is approximately 48 kDa."

9.
What type of trimeric G protein complex would be used by this receptor?
| A. |
| B. |
| C. |
| D. |
| E. |

4. Immunoprecipitation
We did not do any immunoprecipitation experiments, but we did an experiment in which we cultured the cells in the presence of the ligand epinephrine.”
10.
If epinephrine were added to the cells, what would you expect to happen?
| A. |
| B. |
| C. |
| D. |
| E. |

5. Immunofluorescence microscopy
"In the epinephrine cultures, we used a fluorescein-conjugated antibody for immunolocalization of CREB. Here are the data."
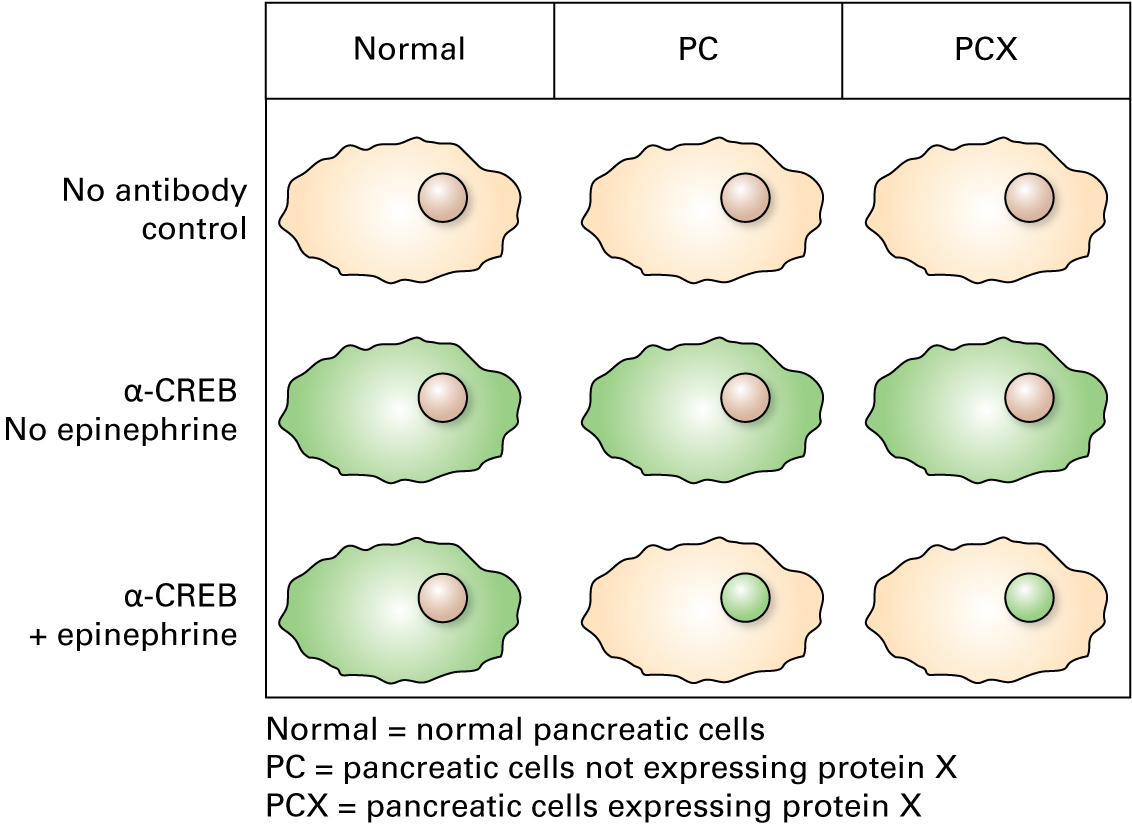
11.
Do these results show what you would expect to see when epinephrine is added to these cells?
| A. |
| B. |
Correct.
Activated transcription factors would be expected to be found in the nucleus.
Incorrect.
Activated transcription factors would be expected to be found in the nucleus.
1.4 Dan's lab: TGF-β signaling pathways
Dan's lab: TGF-β signaling pathways
Kaitlin next stops by Dan's lab. Dan reports:
"Both types of pancreatic cancer cells look like they're expressing TGF-β receptor mRNA, as determined by RT-PCR, and TGF-β receptor protein, as determined by Western blotting. Immunolocalization shows the receptor present on the cell surface. Cells from normal pancreatic tissue don't express TGF-β receptor mRNA or protein."
12.
Which transcription factors should be activated by TGF-β signaling?
| A. |
| B. |
| C. |
| D. |
| E. |
13.
Dan shows Kaitlin the data from cells that were cultured in the presence or absence of TGF-β, then exposed to a fluorescein-conjugated antibody for immunolocalization of Smad:
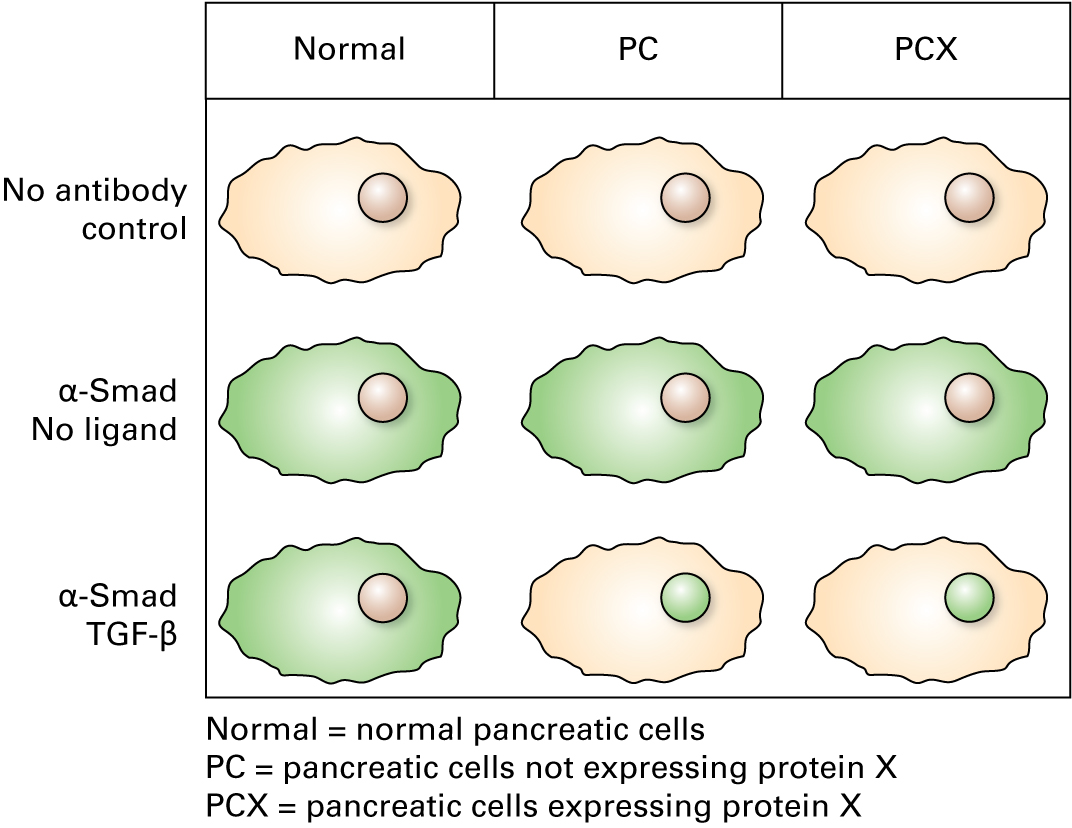
How do you interpret the data?
Adding the ligand TGF-β to cells that express the TGF-β receptor causes the redistribution of the Smad transcription factor to the nucleus, as expected.
1.5 Sarah's lab: Wnt signaling pathways
Sarah's lab: Wnt signaling pathways
Kaitlin next stops by Sarah's lab. Sarah reports:
"We've done characterizations of Frizzled-2 and its co-receptor LRP5 in the different cell lines. Here are the data."
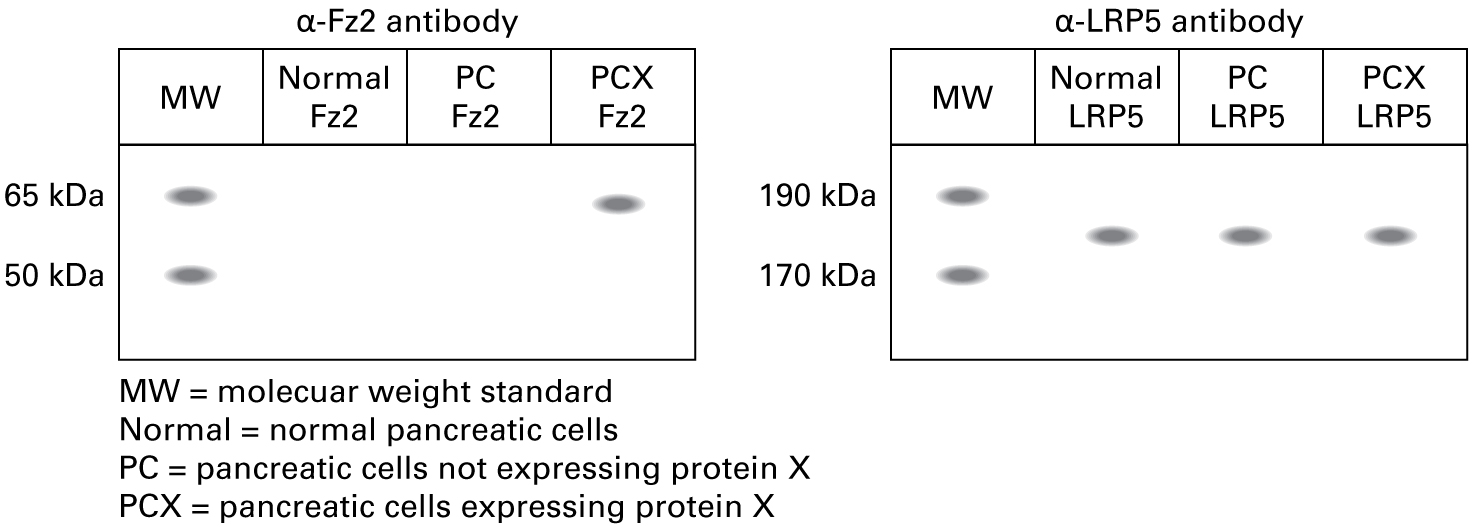
14.
If the ligand Wnt were added to the pancreatic cancer cells expressing Fz2 and LRP5, what would you expect to happen?
| A. |
| B. |
| C. |
| D. |
| E. |
15.
How would you test your answer above?
16.
Performing the immunolocalization experiment gives the results below.
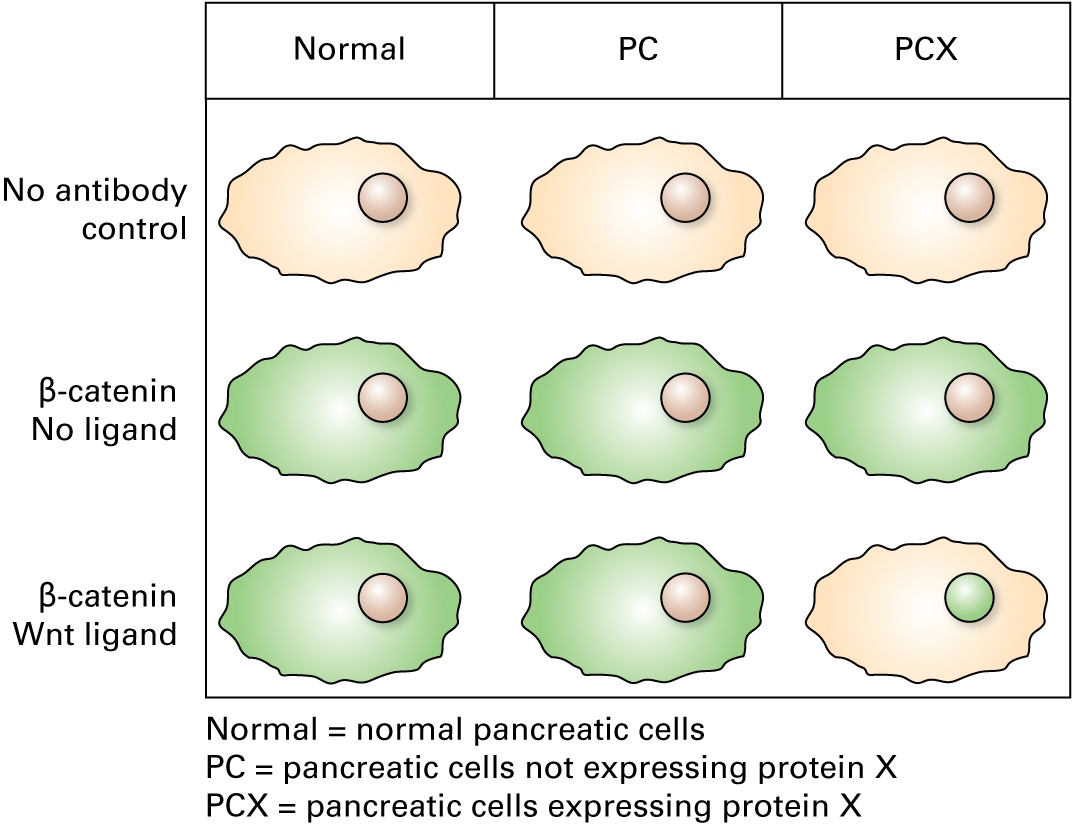
Is this the expected outcome?
| A. |
| B. |
17.
Given what we have learned about the signaling pathways that are activated in the cells, how could you test to see which pathway may be responsible for the expression of protein X?
You could treat cells with the following ligands to test for the induction of protein expression:
- NRG1 for HER2-HER3 signaling
- Epinephrine for β-adrenergic receptor–mediated signaling
- TGF-β for TGF-β receptor–mediated signaling
- Wnt ligand for Fz2-LRP5 signaling
The treated cells could be cultured, and the secreted proteins in the culture medium could be assayed by polyacrylamide gel electrophoresis and Coomassie blue staining to identify a 90-kDa protein product, protein X.
18.
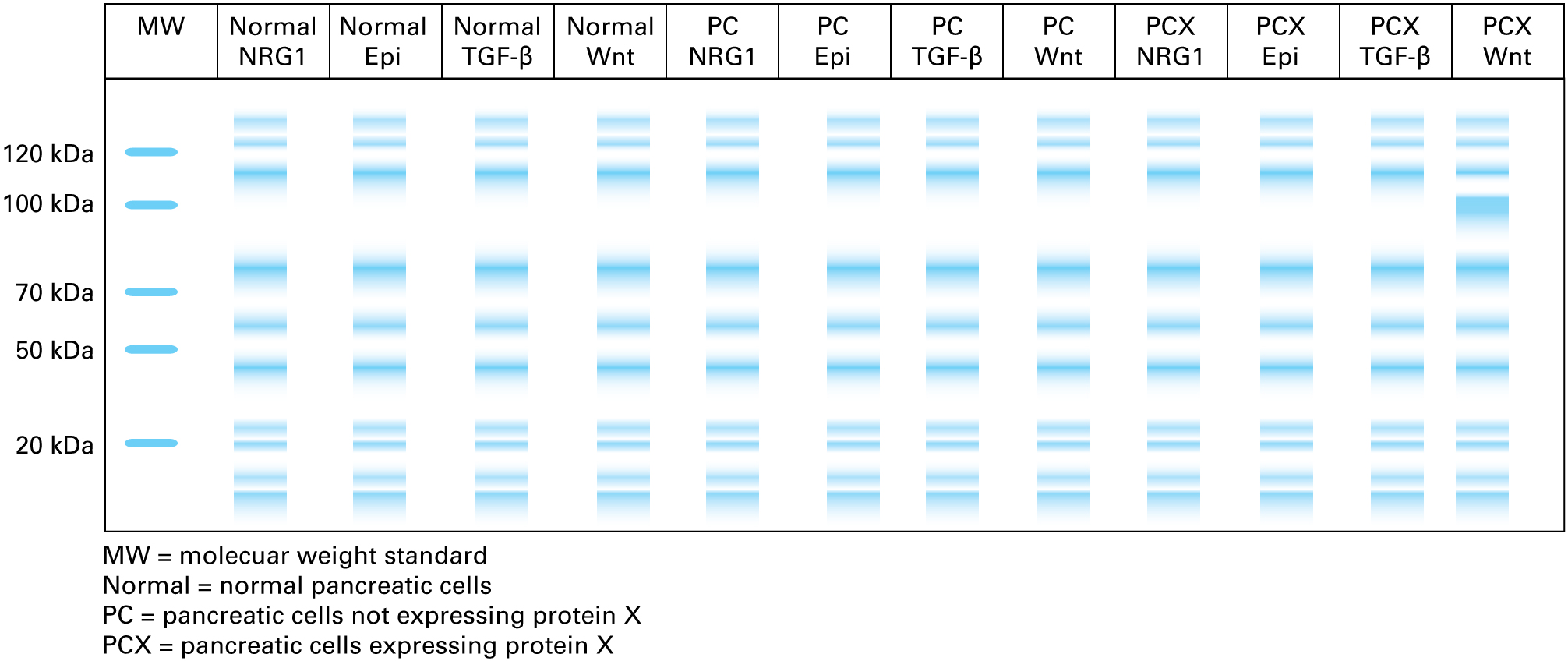
Given the results shown above, which signaling pathway is responsible for the expression of the secreted protein X?
| A. |
| B. |
| C. |
| D. |
| E. |
1.6 Conclusion
Conclusion
19.
What do you think is wrong with the signaling pathways in the aggressive cancer cells?
20.
Sarah sequences the genes for APC, Axin, β-catenin, GSK3, and CK1 from the cancer cell lines and finds DNA mutations that would lead to changes in serine and threonine residues in one of those genes. Which gene do you think is affected?
| A. |
| B. |
| C. |
| D. |
| E. |
Correct.
β-Catenin activity is regulated by phosphorylation at serine and threonine residues through GSK3 and CK1. A reduction or loss in phosphorylation would result in an increased distribution of β-catenin in the nucleus.
Incorrect.
β-Catenin activity is regulated by phosphorylation at serine and threonine residues through GSK3 and CK1. A reduction or loss in phosphorylation would result in an increased distribution of β-catenin in the nucleus.
21.
Protein X is sequenced and identified as NRG2. Which non-Wnt signaling pathway would it affect?
| A. |
| B. |
| C. |
| D. |
| E. |
Correct.
NRG2 would stimulate cell signaling through the HER2-HER3 complex. The aggressive cancer cells appear to have a primary defect in Wnt signaling, which leads to chronic activation of HER2-HER3 signaling.
Incorrect.
NRG2 would stimulate cell signaling through the HER2-HER3 complex. The aggressive cancer cells appear to have a primary defect in Wnt signaling, which leads to chronic activation of HER2-HER3 signaling.
22.
The aggressive pancreatic cancer cells are subsequently shown to have high cytoplasmic levels of SOCS, GRB2, and Sos proteins. Which of the following statements is likely to be true?
| A. |
| B. |
| C. |
| D. |
| E. |
23.
What would be the best approach to treat these cancers?
Activity results are being submitted...
1.7 Additional Information
Additional Information: Stages of FDA Approved Drug Development
FDA Drug Development Process http://www.fda.gov/ForPatients/Approvals/Drugs/default.htm
Step 1: Discovery and Development
Discovery of new drugs can arise from (1) new information about a disease process, (2) testing the therapeutic effects of compounds against many diseases, (3) unexpected effects of current treatments, and (4) new technologies for drug delivery.
Drug development includes determination of (1) mechanisms of absorption, metabolism, and excretion, (2) potential benefits, (3) optimum dosage, (4) optimum delivery, (5) side effects, (6) effects on different groups of people, (7) interactions with other drugs, and (8) comparative effectiveness.
For more information visit: http://www.fda.gov/ForPatients/Approvals/Drugs/ucm405382.htm
Step 2: Preclinical Research
In vitro (cell lines) and in vivo (animal) research is carried out to determine dosing and toxicity levels. Findings are reviewed to determine whether the drug should be tested in humans.
For more information visit: http://www.fda.gov/ForPatients/Approvals/Drugs/ucm405658.htm
Step 3: Clinical Research
The drug is tested in clinical trials in humans. Several factors are important in the design of these clinical trials: (1) selection criteria, (2) number of participants, (3) study length, (4) presence of a control group, (5) drug route and dosage, (6) type of data to be collected, and (7) data analysis plan. Trials consist of four levels, or phases, starting with a small-scale approach and moving to a large-scale approach.
For more information visit: http://www.fda.gov/ForPatients/Approvals/Drugs/ucm405622.htm
Step 4: FDA Review
Drug developers submit data on preclinical and clinical trials and can file an application to market the drug. The FDA reviews the information and decides whether to give approval or not.
For more information visit: http://www.fda.gov/ForPatients/Approvals/Drugs/ucm405570.htm
Step 5: FDA Post-Market Safety Monitoring
The FDA examines problems associated with a drug over its lifetime and can issue warnings or changes depending on the severity of the problems.
For more information visit: http://www.fda.gov/ForPatients/Approvals/Drugs/ucm405579.htm

Your grade of 0% has been submitted.