HOW DO WE KNOW?
FIG. 13.1
How are whole genomes sequenced?
BACKGROUND DNA sequencing technologies can only determine the sequence of DNA fragments far smaller than the genome itself. How can the sequences of these small fragments be used to determine the sequence of an entire genome? In the early years of genome sequencing, many researchers thought that it would be necessary to know first where in the genome each fragment originated before sequencing it. A group at Celera Genomics reasoned that if so many fragments were sequenced that the ends of one would almost always overlap with those of others, then a computer program with sufficient power might be able to assemble the short sequences to reveal the sequence of the entire genome.
HYPOTHESIS A genome sequence can be determined by sequencing small, randomly generated DNA fragments and assembling them into a complete sequence by matching regions of overlap between the fragments.
EXPERIMENT Hundreds of millions of short sequences from the genome of the fruit fly, Drosophila melanogaster, were sequenced. Fig. 13.1a shows examples of overlapping fragments, using a sentence from Watson and Crick’s original paper on the chemical structure of DNA as an analogy.
RESULTS The computer program the group had written to assemble the fragments worked. The researchers were able to sequence the entire Drosophila genome by piecing together the fragments according to their overlaps. In the sentence analogy, the fragments (Fig. 13.1a) can be assembled into the complete sentence (Fig. 13.1b) by matching the overlaps between the fragments.
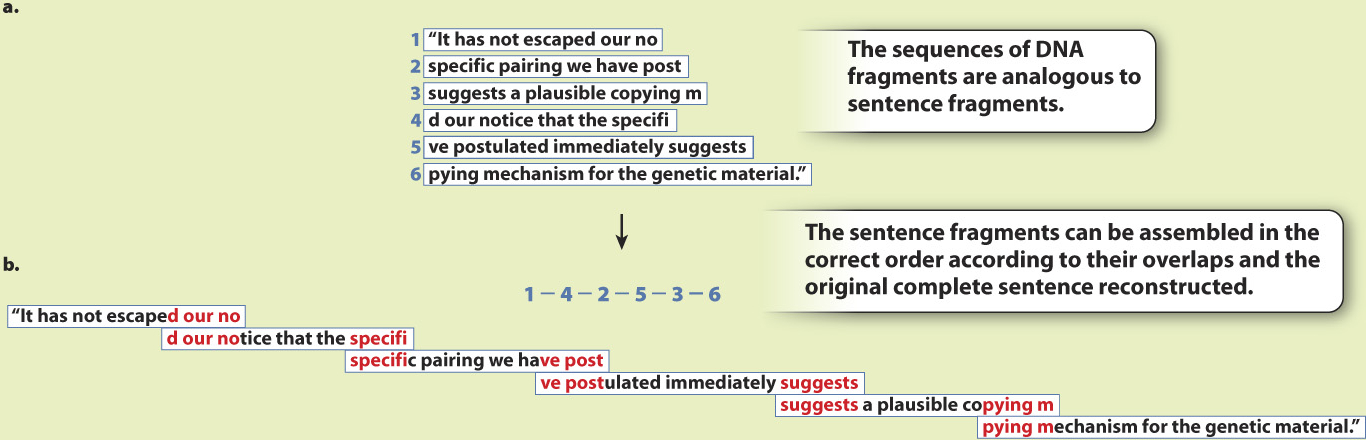
CONCLUSION The hypothesis was supported: Celera Genomics could determine the entire genomic sequence of an organism by sequencing small, random fragments and piecing them together at their overlapping ends.
FOLLOW-
SOURCE Adams, M. D., et al. 2000. “The Genome Sequence of Drosophila melanogaster.” Science 287:2185–