Recombinant DNA combines DNA molecules from two or more sources.
The first application of recombinant DNA technology was the introduction of foreign DNA fragments into the cells of bacteria in the early 1970s. The method is simple and straightforward, and remains one of the mainstays of modern molecular research. It can be used to generate a large quantity of a protein for study or therapeutic use.
The method requires a fragment of double-
A common method for producing recombinant DNA is shown in Fig. 12.19. In order to make sure donor DNA can be fused with vector DNA, both pieces are cut with the same restriction enzyme so they both have the same overhangs. In the example shown in Fig. 12.19, the donor DNA is a fragment produced by digestion of genomic DNA with the restriction enzyme EcoRI, resulting in four-
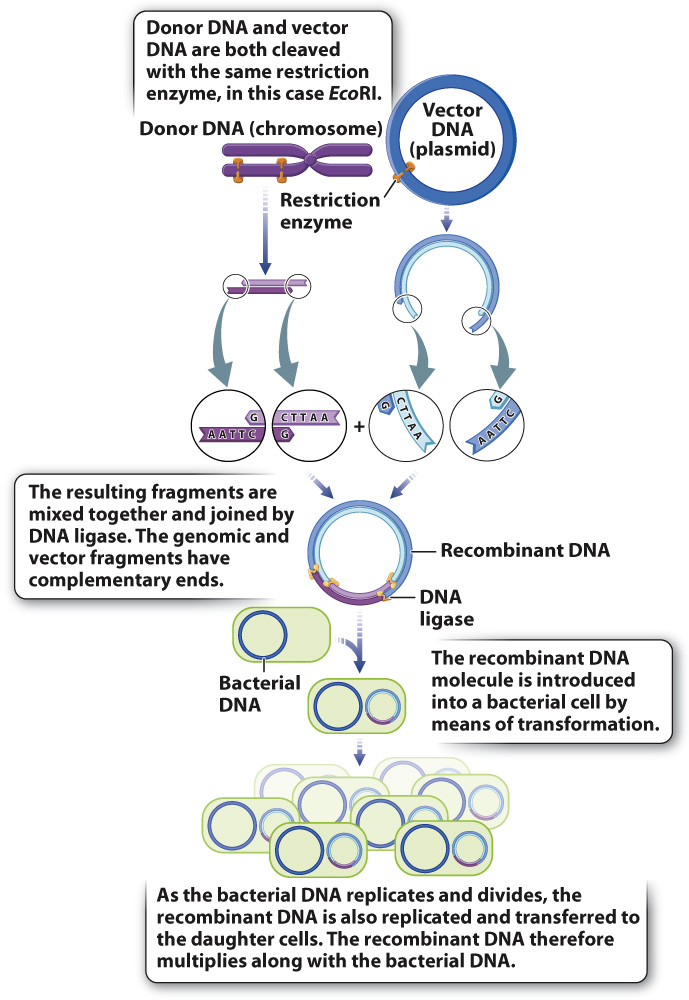
The next step in the procedure is transformation, in which the recombinant DNA is mixed with bacteria that have been chemically coaxed into a physiological state in which they take up DNA from outside the cell. Having taken up the recombinant DNA, the bacterial cells are transferred into growth medium, where they multiply. Since the vector part of the recombinant DNA molecule contains all the DNA sequences needed for its replication and partition into the daughter cells, the recombinant DNA multiplies as the bacterial cell multiplies. If the recombinant DNA functions inside the bacterial cell, then new genetic characteristics may be expressed by the bacteria. For example, the recombinant DNA may allow the bacterial cells to produce a human protein, such as insulin or growth hormone.
Quick Check 4 In making recombinant DNA molecules that combine restriction fragments from different organisms, researchers usually prefer restriction enzymes like BamHI or HindIII that generate fragments with “sticky ends” (ends with overhangs) rather than enzymes like HpaI or SmaI (Table 12.1) that generate fragments with “blunt ends” (ends without overhangs). Can you think of a reason for this preference?
Quick Check 4 Answer
The value of sticky ends is that they give the researcher greater control over which restriction fragments can come together and be attached. Sticky ends can pair only with other sticky ends that have complementary 3′ and 5′ overhangs. Hence, restriction fragments produced by BamHI can combine only with other fragments produced by BamHI, and not with fragments produced by, for example, HindIII. However, any blunt end can be attached to any other blunt end, for example a HpaI end to a SmaI end.