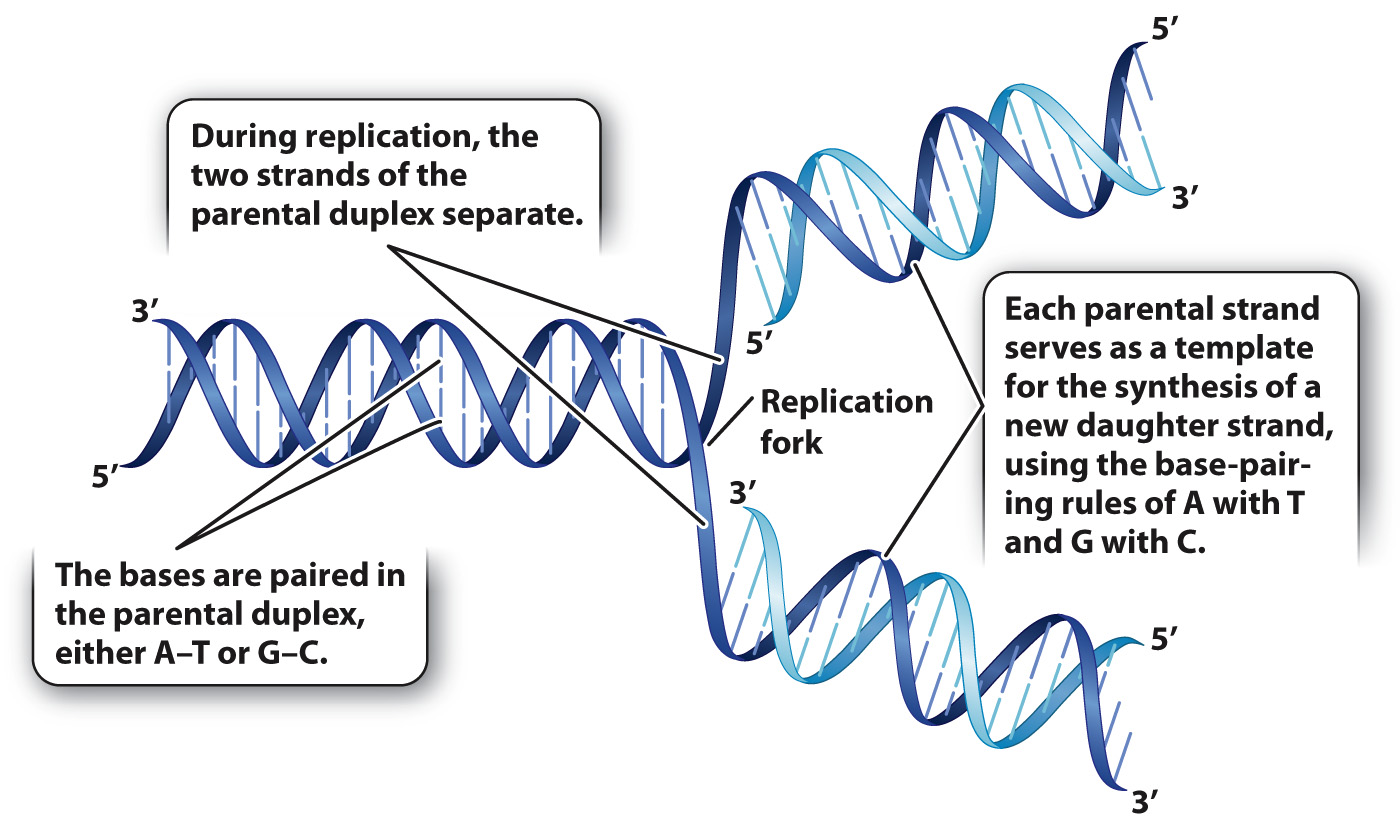
During DNA replication, the parental strands separate and new partners are made.
When Watson and Crick published their paper describing the structure of DNA, they also coyly laid claim to another discovery: “It has not escaped our notice that the specific pairing we have postulated [A with T, and G with C] immediately suggests a copying mechanism for the genetic material.” The copying mechanism they had in mind is exquisitely simple. The two strands of the parental duplex molecule separate (Fig. 12.1), and each individual parental strand serves as a model, or template strand, for the synthesis of a daughter strand. As each daughter strand is synthesized, the order of the bases in the template strand determines the order of the complementary bases added to the daughter strand. For example, the sequence 5′-ATGC-
A key prediction of the model shown in Fig. 12.1 is semiconservative replication. That is, after replication, each new DNA duplex consists of one strand that was originally part of the parental duplex and one newly synthesized strand. An alternative model is conservative replication, which proposes that the original DNA duplex remains intact and the daughter DNA duplex is completely new. Which model is correct? If there were a way to distinguish newly synthesized daughter DNA strands (“new strands”) from previously synthesized parental strands (“old strands”), the products of replication could be observed and the mode of replication determined.
American molecular biologists Matthew S. Meselson and Franklin W. Stahl carried out an experiment to determine how DNA replicates. This experiment, described in Fig. 12.2, has been called “the most beautiful experiment in biology” because it so elegantly demonstrated the scientific method (Chapter 1) of hypothesis, prediction, and experimental test. They distinguished “old” from “new” DNA strands by labeling them with nonradioactive isotopes of nitrogen that have different densities: the normal form of nitrogen, 14N, and a heavier form with an extra neutron, denoted 15N.
Meselson and Stahl found that DNA in fact replicates semiconservatively (Fig. 12.2). This finding also predicted the results when cells are allowed to undergo two rounds of replication in a medium containing only light 14N nitrogen. The heavy strand and light strand each serve as templates for a new light daughter strand. The result is that half of the DNA molecules will have one heavy old strand and one light new strand and an intermediate density, and half of the DNA molecules will have one light old strand and one light new strand and a low density. This is precisely what they observed (Fig. 12.2).
HOW DO WE KNOW?
FIG. 12.2
How is DNA replicated?
BACKGROUND Watson and Crick’s discovery of the structure of DNA in 1953 suggested a mechanism by which DNA is replicated. Experimental evidence came from research by American molecular biologists Matthew Meselson and Franklin Stahl in 1958.
HYPOTHESIS DNA replicates in a semiconservative manner, meaning that each new DNA molecule consists of one parental strand and one newly synthesized strand.
ALTERNATIVE HYPOTHESIS DNA replicates in a conservative manner, meaning that one DNA molecule consists of two parental strands, and the other consists of two newly synthesized strands.
METHOD Meselson and Stahl distinguished parental strands (“old”) from newly synthesized strands (“new”) using two isotopes of nitrogen atoms. Old strands were labeled with a heavy form of nitrogen with an extra neutron (15N), and new strands were labeled with the normal, lighter form of nitrogen (14N).
EXPERIMENT The researchers first grew bacterial cells on medium containing only the heavy 15N form of nitrogen. As the cells grew, 15N was incorporated into the DNA bases, resulting, after several generations, in DNA containing only 15N. They then transferred the cells into medium containing only light 14N nitrogen. After one round of replication in this medium, cell replication was halted. The researchers could not observe the DNA directly, but instead they measured the density of the DNA by spinning it in a high-
PREDICTION If DNA replicates conservatively, half of the DNA in the cells should be composed of two heavy parental strands containing15N, and half should be composed of two light daughter strands containing 14N. If DNA replicates semiconservatively, the daughter DNA molecules should each consist of one heavy strand and one light strand.
RESULTS When the fully15N-
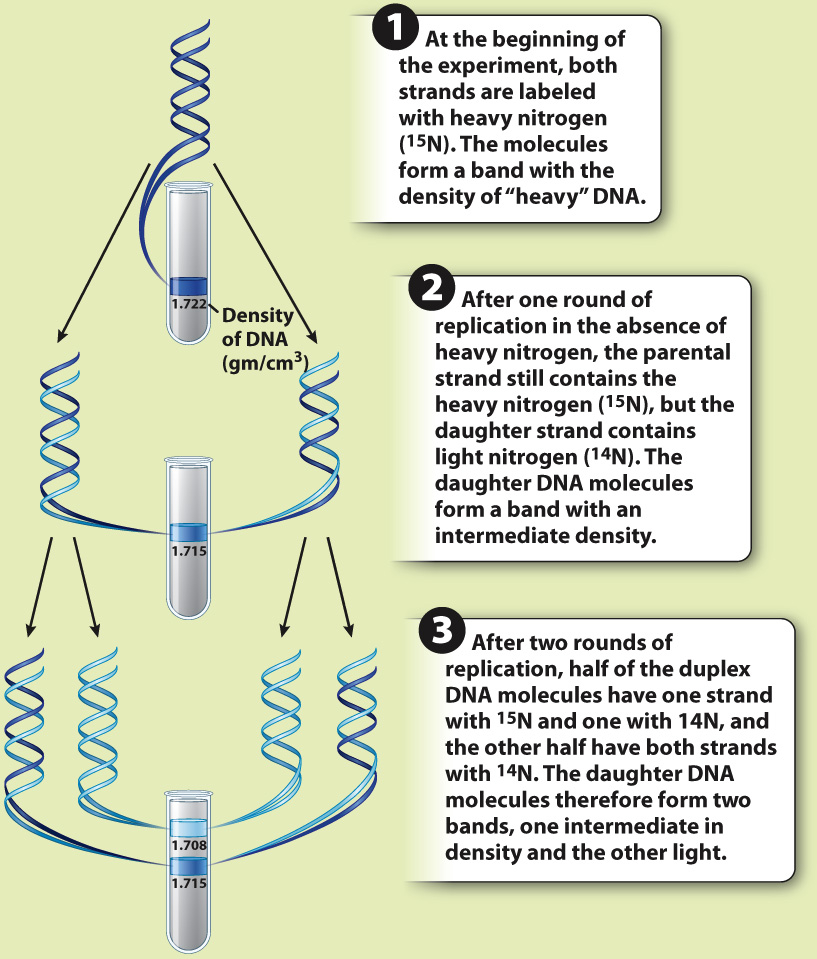
CONCLUSION DNA replicates semiconservatively, supporting the first hypothesis.
SOURCE Meselson, M., and F. W. Stahl. 1958. “The Replication of DNA in Escherichia coli.” PNAS 44:671–
Quick Check 1 Suppose Meselson and Stahl had done their experiment the other way around, starting with cells fully labeled with 14N light DNA and then transferring them to medium containing only 15N heavy DNA. What density of DNA molecule would you predict after one and two rounds of replication?
Quick Check 1 Answer
After one round of replication, you would predict only 14N/15N hybrid DNA, which has a density of 1.715 gm/cm3. After two rounds of replication, you would predict half the molecules to be 14N/15N hybrid DNA (density 1.715 gm/cm3) and half to be 15N/15N heavy DNA (density 1.722 gm/cm3).
Important as it was in demonstrating semiconservative replication in bacteria, the Meselson–