Chapter 13 Summary
Core Concepts Summary
13.1 A genome is the genetic material of a cell, organism, organelle, or virus, and its sequence is the order of bases along the DNA or (in some viruses) RNA.
The sequence of an organism’s genome can be determined by breaking up the genome into small fragments, sequencing these fragments, and then putting the sequences together at their overlaps. page 272
Sequences that are repeated in the genome can make sequence assembly difficult. page 273
13.2 Researchers annotate genome sequences to identify genes and other functional elements.
Genome annotation is the process by which the types and locations of the different kinds of sequences, such as protein-
Genome annotation sometimes involves scanning the DNA sequence for characteristic sequence motifs. page 276
Comparison of DNA sequences with messenger RNA sequences reveals the intron–
By comparing annotated genomes of different organisms, we can gain insight into their ancestry and evolution. page 277
The annotated HIV genome shows it is a retrovirus, and it contains the genes gag, pol, and env. page 277
13.3 The number of genes in a genome and the size of a genome do not correlate well with the complexity of an organism.
The C-
In eukaryotes, the C-
13.4 The orderly packaging of DNA allows it to carry out its functions and fit inside the cell.
Bacteria package their circular DNA in a structure called a nucleoid. page 281
Eukaryotic cells package their DNA into linear chromosomes. page 282
DNA in eukaryotes is wound around groups of histone proteins called nucleosomes to form a 10-
Diploid organisms have two copies of each chromosome, called homologous chromosomes. page 284
Humans have 23 pairs of chromosomes, including the X and Y sex-
The genomes of mitochondria and chloroplasts are organized into nucleoids that resemble, but are distinct from, those of bacteria. page 285
13.5 Viruses have diverse genomes, but all require a host cell to replicate.
Viruses can be classified by the Baltimore system, which defines seven groups on the basis of type of nucleic acid and the way the mRNA is synthesized. page 286
Viruses can infect all types of organism, but a given virus can infect only some types of cell. page 287
The host range of a virus is determined by proteins on viral and host cells. page 287
Viruses have diverse shapes, including head-
Viruses are capable of molecular self-
Self-Assessment
Describe the shotgun method for determining the complete genome sequence of an organism.
Self-Assessment 1 Answer
Shotgun sequencing is an approach for determining the complete genome sequence of an organism. It involves first chopping up the chromosome or genome into pieces small enough to be sequenced by current sequencing technology. These small pieces are then sequenced 10–50 times, to reduce the chance of making an error, and then assembled, mainly through computer programs, according to their overlaps. The result is the long, continuous sequence of nucleotides in the chromosome or genome.
Repeated sequences can be classified according to their organization in the genome as well as according to their function. Give at least two examples of each.
Self-Assessment 2 Answer
Repeated sequences classified according to their organization in the genome include dispersed repeats (a repeated nucleotide sequence that is dispersed throughout the DNA), tandem repeats (a nucleotide sequence that is repeated one after another in the DNA), and simple-
sequence repeats (a nucleotide sequence shorter than a tandem repeat that is repeated one after another in the DNA). Repeated sequences classified according to their function include DNA transposons, retrotransposons, and alpha satellites, as well as repeated sequences that fall into none of these categories, like nonfunctional gene duplicates. Explain the purpose of genome annotation.
Self-Assessment 3 Answer
The purpose of genome annotation is to determine the function of various sequences on the genome. Scientists can then summarize the knowledge found in the genome by comparing similar motifs between organisms, to guide research, and to reveal evolutionary relationships among organisms. Genome annotation allows scientists to make an educated guess, or hypothesis, concerning a novel gene based on its similarity to known genes.
Describe how the comparison of genomic DNA to messenger RNA can identify the exons and introns in a gene.
Self-Assessment 4 Answer
In the process of mRNA maturing, the introns are spliced out of the mRNA sequence, typically leaving only the exons behind. If you were to then compare the mRNA sequence to the DNA from which it was transcribed, you could see which DNA sequences were complementary to the exons and then, based on the sequences that are missing in the mRNA, which sequences were complementary to introns.
Explain how comparing the sequences of two genomes can help to infer evolutionary relationships.
Self-Assessment 5 Answer
Sequences that are similar in different organisms are said to be conserved and usually have an important function. By looking at a conserved gene in different species, scientists can determine an evolutionary timeline of the slight changes seen in that gene and map it to the organisms that have the gene, inferring relationships among them.
- Page 290
What are some reasons why, in multicellular eukaryotes, genome size is not necessarily related to number of protein-
coding genes or organismal complexity? Self-Assessment 6 Answer
The genomes of multicellular eukaryotes can have vastly different amounts of repetitive sequences, including dispersed repeats, tandem repeats, and transposable elements of various kinds. The abundance of repetitive sequences has no relation to the number of protein-
coding genes or organismal complexity, but it does affect genome size. Hence, genome size is not necessarily related to the number of protein- coding genes or organismal complexity. The disconnect between genome size and organismal complexity in multicellular eukaryotes is known as the C- value paradox. Compare and contrast the mechanisms by which bacterial cells and eukaryotic cells package their DNA.
Self-Assessment 7 Answer
In bacterial cells, DNA is packaged through the activity of the enzyme topoisomerase II and the formation of supercoils. Topoisomerase II breaks the double helix of the DNA, rotates the ends, and then seals the break. This results in underwinding, which creates strain on the DNA molecule. This strain is relieved by the formation of supercoils in which the DNA molecule coils on itself. These supercoils then form a structure with multiple loops, bound by proteins, called the nucleoid. Eukaryotic cells, in contrast, wind their DNA around a group of histone proteins called a nucleosome. The histone proteins are positively charged, thus balancing the negatively charged DNA backbone. The nucleosomes and DNA are then coiled to form the 30-
nm chromatin fiber. Draw a nucleosome, indicating the positions of DNA and proteins.
Self-Assessment 8 Answer
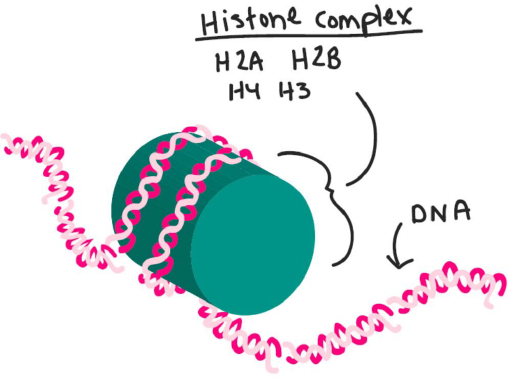
Define “homologous chromosomes” and describe a technique that you could use to show their similarity.
Self-Assessment 9 Answer
Homologous chromosomes are pairs of chromosomes that match in size and appearance. DNA sequencing demonstrates that they have the same genes (not necessarily identical in sequence) arranged in the same order. Cytologically, chromosome painting (the labeling of DNA with complementary sequences that have different fluorescent dyes attached to them) can show the similarity between homologous chromosomes because, under fluorescent light, the homologous chromosomes fluoresce at the same wavelength.
Describe the steps necessary to synthesize mRNA from each of the following: double-
stranded DNA, single- stranded (+)DNA, single- stranded (–)DNA, single- stranded (+)RNA, and single- stranded (–)RNA. Self-Assessment 10 Answer
Double-
stranded DNA → mRNA Single-
stranded (+)DNA → double- stranded DNA → mRNA Single-
stranded (–)DNA → double- stranded DNA → mRNA Single-
stranded (+)RNA → single- stranded –DNA → double- stranded DNA → mRNA Single-
stranded (+)RNA → single- stranded –RNA → mRNA Single-
stranded (–)RNA → mRNA