Some polymorphisms add or remove restriction sites in the DNA.
Restriction enzymes, which cleave double-
Fig. 15.6 shows an example. Here, a restriction enzyme cleaves DNA near the sequence GAGGAG. The three sites shown are in and flanking the β-globin gene from the sickle-
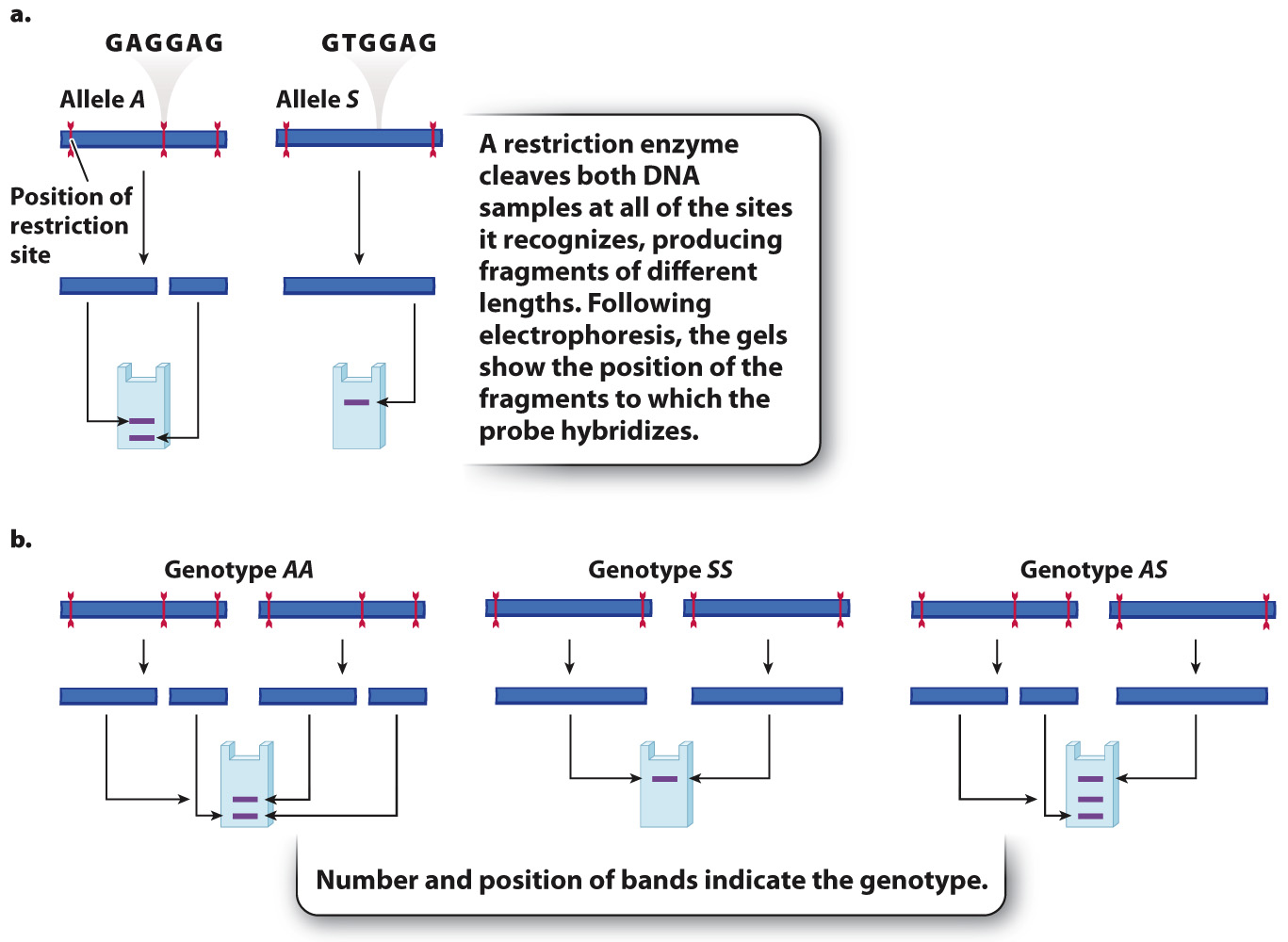
These differences can be visualized by gel electrophoresis (Chapter 12), in which DNA is isolated from different individuals, cut with restriction enzymes, and separated by size on a gel. In this case, we use a restriction enzyme that recognizes the GAGGAG sequence. The A allele produces two fragments: a short fragment that moves rapidly through the gel and ends up near the bottom, and a longer fragment that ends up near the middle. The fragment produced by the S allele is even longer because there is no restriction site in the middle. This fragment ends up forming a band near the top of the gel.
Just as with VNTRs, any individual can carry at most two different alleles of a restriction fragment length polymorphism. As seen in the RFLP in Fig. 15.6b, DNA from homozygous AA individuals yields only the short and medium fragments, and that from homozygous SS individuals yields only the long fragment. DNA from heterozygous AS individuals yields all three fragments. Therefore, each of the three genotypes AA, AS, and SS yields a unique pattern of bands, allowing each of the possible genotypes to be identified.
How many restriction fragment length polymorphisms are there in the human genome? As in the case of VNTRs, the human genome has many restriction sites, so there are numerous RFLPs in the human population, making it another type of polymorphism that can be used in DNA typing.
Quick Check 4 When a segment of DNA containing either a VNTR or an RFLP is analyzed, the result is fragments of DNA of different lengths. How, then, are VNTRs and RFLPs different?
Quick Check 4 Answer
In a VNTR, the restriction fragments are different lengths because of a variable number of repeats between restriction sites (one, two, three, four, or more). In a RFLP, the restriction fragments are different lengths because a restriction site is removed in one DNA sequence and not in another, making the distances between restriction sites different.