Areas of the genome with variable numbers of tandem repeats are useful in DNA typing.
At many locations in the human genome, short (10–
Any of several methods can be used to DNA fingerprint someone on the basis of that individual’s VNTRs. Amplification by the polymerase chain reaction (PCR, Chapter 12) is a convenient and reliable technique because each VNTR in the genome is flanked by a unique sequence that can be used to design PCR primers that will amplify one and only one VNTR. Although the number of alleles of a particular VNTR can be very large, an example with only five alleles is shown in Fig. 15.5, where the repeated sequence is shown in green between the flanking unique sequences.
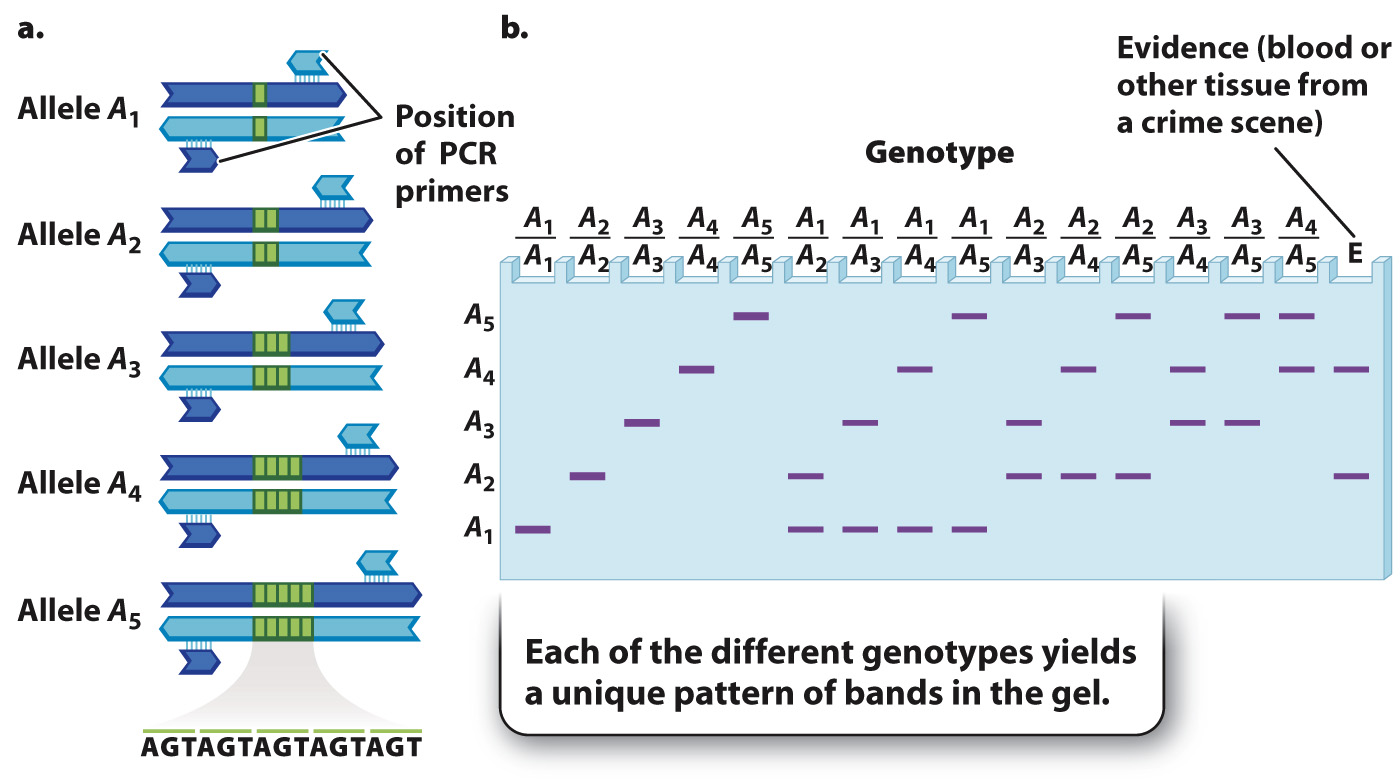
No matter how many alleles exist in the population as a whole, any one diploid individual can have only two alleles, which are present at the same positions in the homologous chromosomes inherited from the mother and father. With five alleles there are 5 possible homozygous genotypes and 10 possible heterozygous genotypes. These can be visualized after PCR by separating the resulting molecules by size by gel electrophoresis (Chapter 12). The pattern of bands expected from the DNA in each genotype formed from the alleles in Fig. 15.5a is shown in Fig. 15.5b. Note that each genotype yields a distinct pattern of bands, and so these patterns can be used to identify the genotype of any individual for a particular region of DNA.
With as few as 10 equally frequent alleles, the likelihood that any two individuals would have the same genotype at a particular location by chance alone is about 1 in 50, and with 20 equally frequent alleles the likelihood decreases to about 1 in 200. These numbers suggest why DNA typing can yield a genetic fingerprint that can uniquely match DNA samples.
The lane labeled “E” at the far right in Fig. 15.5b shows the DNA bands for this same polymorphism from biological material collected at the scene of a crime. In this case, the bands in lane E imply that the source of the evidence is an individual of genotype A2/A4 because this is the only genotype with bands with vertical positions in the gel that exactly match those of the evidence sample. A single polymorphism of the type shown in Fig. 15.5 is not enough to establish for sure that two pieces of DNA come from the same person, but because the human genome has polymorphic sites scattered throughout, many polymorphisms can be examined. If six or eight of these polymorphisms match between two samples, it is very likely that the samples come from the same individual (or from identical twins). On the other hand, if any of the polymorphisms fails to match band for band, then it is almost certain that the samples come from different individuals (barring such technical mishaps as samples whose DNA is contaminated, degraded, or mislabeled).
One of the advantages of DNA typing is that a large amount of DNA is not needed. Even minuscule amounts of DNA can be typed, so tiny spots of blood, semen, or saliva are sufficient samples. A cotton swab wiped across the inside of your cheek will contain enough DNA for typing; so will a discarded paper cup or a cigarette butt. One enterprising researcher in England used PCR to type the dog feces on his lawn and matched it to the DNA from hair he obtained from neighborhood dogs, much to the chagrin of the guilty dog’s owner.
Quick Check 3 In typing DNA from a sample found at a crime scene, how can a DNA mismatch prove that a suspect is not the source of that sample, whereas a DNA match does not necessarily prove that a suspect is the source?
Quick Check 3 Answer
If the suspect is the actual source of a sample, then the DNA fingerprints must match exactly, and any mismatch rules out the suspect as the source of the sample. When there is a match, however, there is always a small chance (typically very small) that the sample came from another person with the same DNA fingerprint.