Genetic risk factors for disease can be localized by genetic mapping.
The discovery of abundant genetic variation in DNA sequences in human populations, such as single-nucleotide polymorphisms (Chapter 15), made it possible to study genetic linkage in the human genome. At first, the focus was on finding mutations that cause disease, such as the mutation that causes cystic fibrosis (Chapter 14). The method was to study large families extending over three or more generations in which the disease was present and then to identify the genotypes of each of the individuals for thousands of genetic markers (previously discovered DNA polymorphisms) throughout the genome. The goal was to find genetic markers that showed a statistical association with the disease gene, which would indicate genetic linkage and reveal the approximate location of the disease gene along the chromosome.
Page 357
Fig. 17.12 illustrates the underlying concept. It assumes 100 chromosomes observed among different individuals in a pedigree, of which 50 carry a mutant allele of a gene and 50 carry a nonmutant allele. These chromosomes are tested for a marker of known location, in this case a single-nucleotide polymorphism (SNP) in which one of the nucleotide pairs in the DNA is a G–C base pair in some chromosomes and A–T in others. In Fig. 17.12a, there is clearly an association between the disease gene and the SNP. Almost all of the chromosomes that carry the mutant allele show the G–C nucleotide pair, whereas almost all of the chromosomes that carry the nonmutant allele show the A–T nucleotide pair. The two chromosomes in which the mutant and nonmutant genes are associated with the other SNPs can be attributed to recombination.
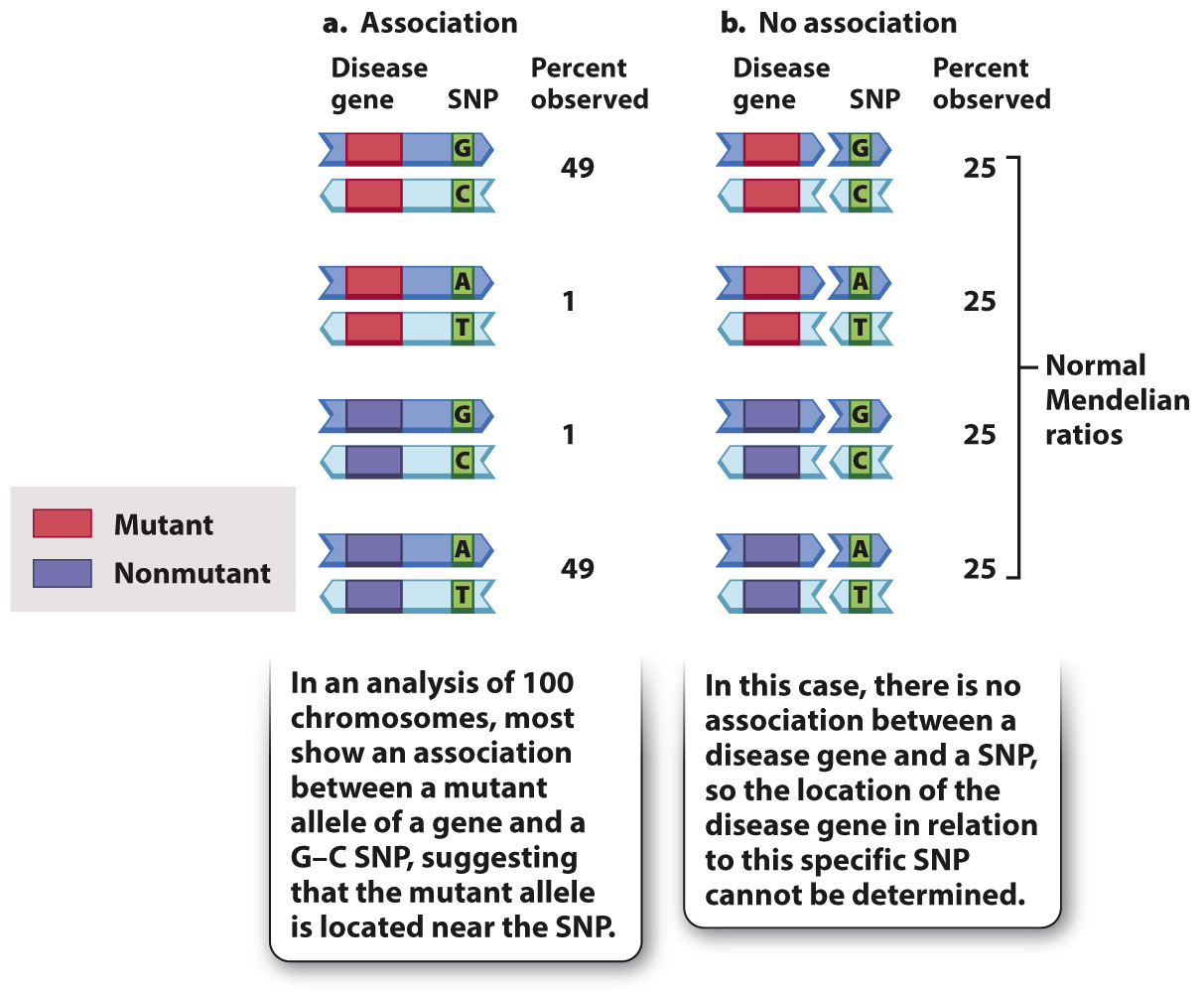
FIG. 17.12 Genetic mapping. SNPs associated with a mutant gene show where that gene is located in the genetic map.
The association in Fig. 17.12a may be contrasted with the pattern in Fig. 17.12b, in which there is no association. In this case, each of the alleles of the disease gene is equally likely to carry either form of the SNP. The failure to find an association means that the SNP is not closely linked to the disease gene, and may in fact be in a different chromosome. In actual studies, an association is almost never observed, but the lucky find of an association helps identify the location of the disease gene in the genetic map. Using such association methods, hundreds of important disease genes have been located by genetic mapping. Once the location of the disease gene is known, the identity and normal function of the gene can be determined.
