The identity of the floral organs is determined by combinatorial control.
The genetic control of flower development in Arabidopsis was discovered by the analysis of mutant plants, an investigation similar in principle to the way that genetic control of development in fruit flies was determined. The plant mutants fell into three classes showing characteristic floral abnormalities (Fig. 20.20). Mutations in the gene apetala-
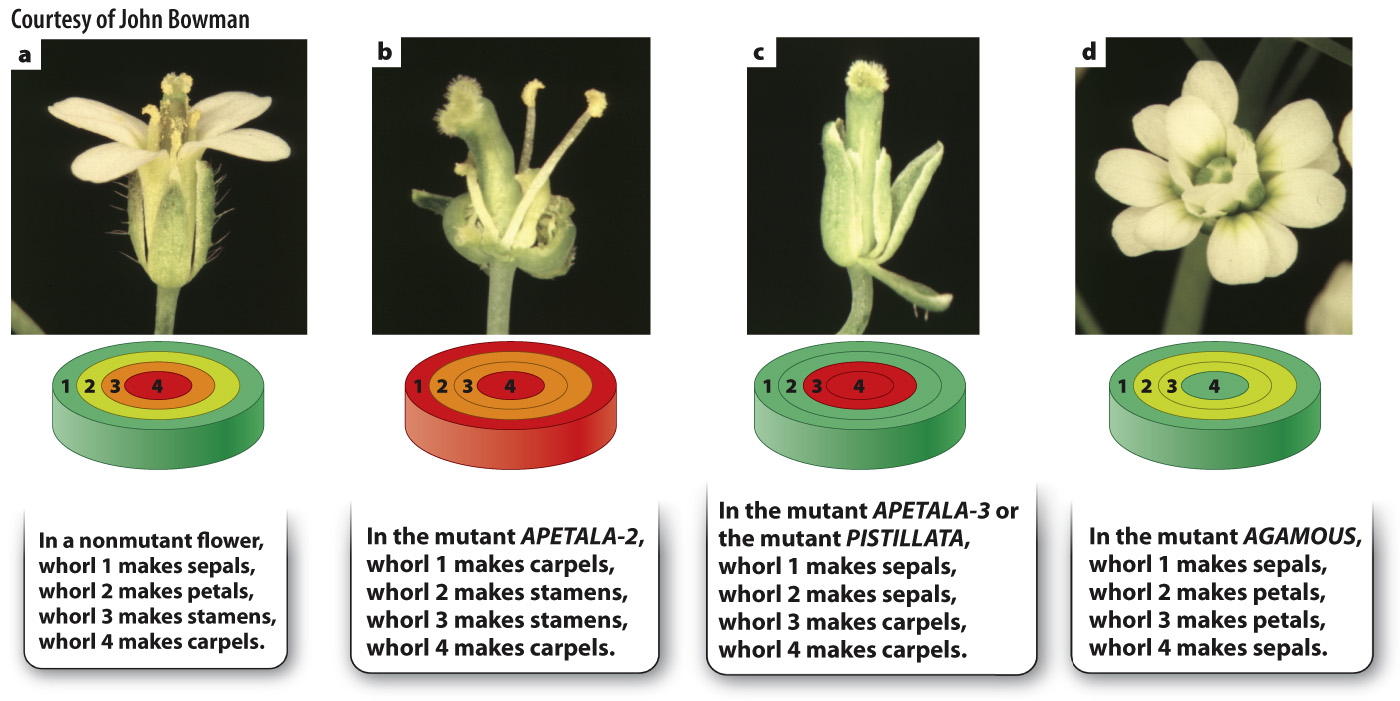
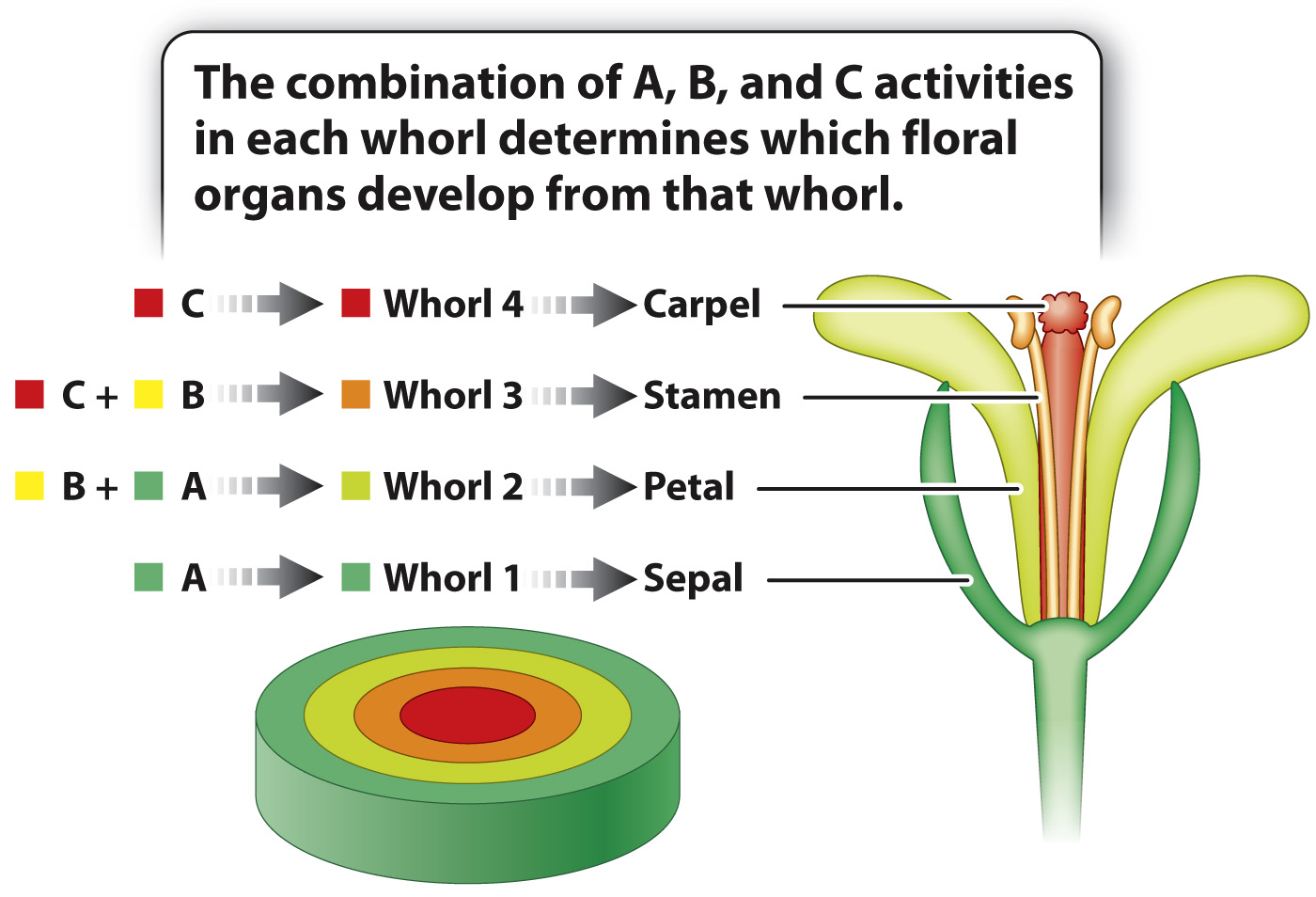
The observation that the mutant phenotypes affect development of pairs of adjacent whorls already hints at combinatorial control, which researchers incorporated into a model of floral development called the ABC model. The model invokes three activities arbitrarily called A, B, and C. These activities represent the function of one or more different proteins that are hypothesized to be present in cells of each whorl as shown in Fig. 20.21. In the course of floral differentiation, the cells of each whorl become different from those in the other whorls, and different combinations of genes are activated or repressed in each whorl. Activity A is present in whorls 1 and 2, activity B in whorls 2 and 3, and activity C in whorls 3 and 4. According to the ABC model, activity A alone results in the formation of sepals, A and B together result in petals, B and C together result in stamens, and C alone results in carpels. Like any good hypothesis (Chapter 1), this one makes specific predictions. Mutants lacking A will have defects in whorls 1 and 2, mutants lacking B will have defects in whorls 2 and 3, and mutants lacking C will have defects in whorls 3 and 4.
Matching these predictions with the mutants in Fig. 20.20 yields the following correspondence:
The A activity is encoded by the gene APETALA-
2. The B activity is encoded jointly by the genes APETALA-
3 and PISTILLATA.The C activity is encoded by the gene AGAMOUS.
To account for the mutant phenotypes in Fig. 20.20, you need only to postulate that when A is absent, C is expressed in whorls 1 and 2 in addition to its normal expression in whorls 3 and 4; and when C is absent, A expression expands into whorls 3 and 4 in addition to its normal expression in whorls 1 and 2. The domains of expression expand in this way because, in addition to its role in activating certain downstream genes, the product of APETALA-
Quick Check 6 Predict the phenotype of a flower in which APETALA-
Quick Check 6 Answer
Because A and B together result in petals, and B and C together result in stamens, a flower expressing B activity in all four whorls would have, from outside to inside, petals, petals, stamens, and stamens.
The identification of the products of these genes demonstrates combinatorial control and explains the mutant phenotypes. All of the genes encode transcription factors, which are called AP2, AP3, PI, and AG, corresponding to the four genes. The transcription factors bind to cis-
Although other transcription factors associated with other activities that also contribute to flower development were discovered later, the original ABC model is still valid and serves as an elegant example of combinatorial control. As in the case of the Pax6 gene in the development of animals’ eyes, evolution of the cis-
