26.4 The Diversity of Bacteria
Traditionally, bacterial taxonomy was a laborious process. Microbiologists grew bacteria in beakers or on petri dishes, carefully separating and enriching individual populations until only one type of bacterium—
With molecular sequencing technologies, however, bacterial species are now characterized by their DNA sequences, especially the genes for the small subunit of ribosomal RNA (rRNA). A novel application of sequencing technology paved the way for modern studies of bacterial diversity and ecology. Microbiologists reasoned that the same procedures used to extract, amplify, and sequence genes from pure cultures in the lab could be used to study bacteria in nature. That is, extracting and reading gene sequences directly from soil or seawater could reveal the diversity of the microbial communities in these environments.
The results were stunning. Most bacteria in nature—
More recently, the application of shotgun sequencing techniques (Chapter 13) to the environmental sampling of genomes has allowed microbiologists to begin matching uncultured populations with specific metabolisms. That is, we need no longer be content to know from molecular sequences what is living in a given environment. Now we can begin to understand what the different microorganisms are doing there. For this reason, another revolution in our understanding of the microbial world lies just around the corner (Fig. 26.13).
HOW DO WE KNOW?
FIG. 26.13
How many kinds of bacterium live in the oceans?
BACKGROUND The oceans are full of bacteria—
METHOD 1 Different types of bacterium can be recognized by the unique nucleotide sequences of their genes. In fact, individual types of bacterium can be identified on the basis of nucleotide sequence in a relatively small region of the gene for the small subunit of ribosomal RNA (rRNA). This molecular “barcode” can be sequenced quickly and accurately for samples of DNA drawn from the environment.
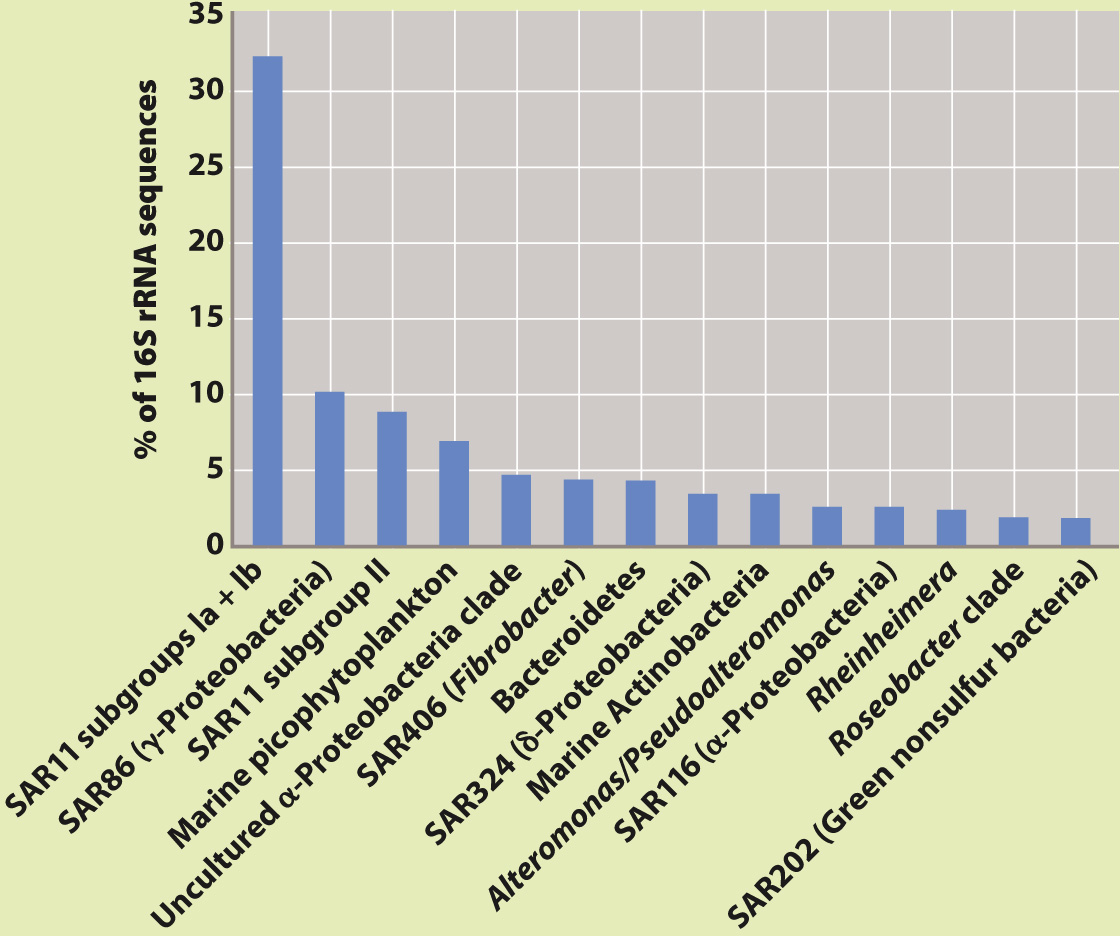
Using this strategy, scientists associated with the International Census of Marine Life obtained samples of seawater and seafloor sediment from deep-
METHOD 2 In another set of experiments, scientists collected samples of seawater from the Sargasso Sea, and from these they amplified whole genomes—
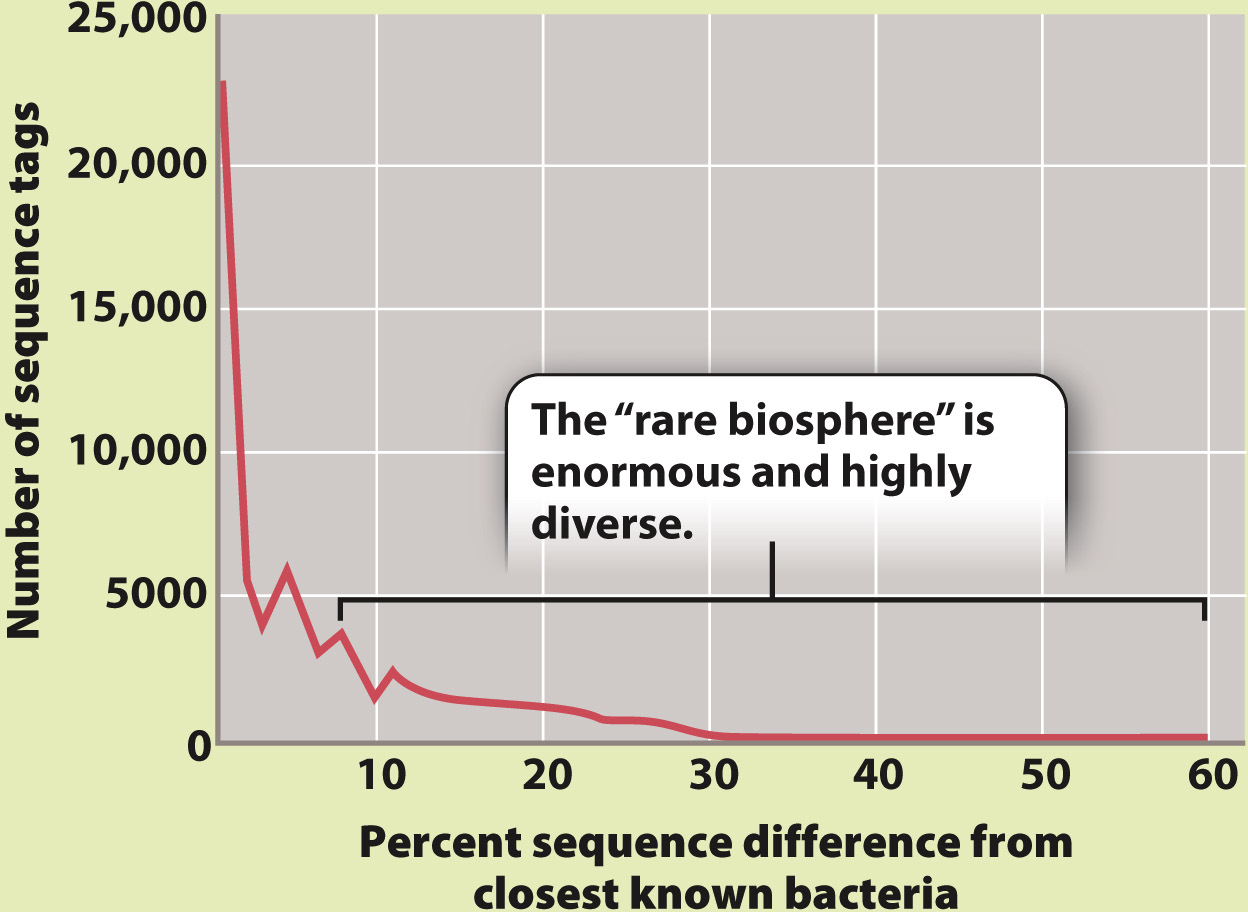
RESULTS Both surveys found that the bacterial diversity of marine environments is much higher than had been thought. The barcode survey (method 1) found as many as 20,000 distinct types of bacteria in a single liter of seawater and estimated that as many as 5–
CONCLUSION The bacterial diversity of the oceans is huge and still largely unexplored. Both barcode and genomic surveys are continuing, and promise a more nearly complete accounting of marine diversity. Together, these surveys will help us understand how marine bacteria function and how they sort into communities.
SOURCES Venter, J. C., et al. 2004. “Environmental Genome Shotgun Sequencing of the Sargasso Sea.” Science 304:66–