Genomic rearrangement generates antibody diversity.
We pointed out earlier that antibody specificity is determined before exposure to the antigen. Clonal selection therefore requires a mechanism for producing a great diversity of antibodies. How is this diversity achieved? Diversity would result if each antibody were encoded by a separate gene. However, this cannot be the case, as the number of antibodies produced by an organism far exceeds the total number of genes in the genome. For example, the human genome consists of about 25,000 protein-
In 1965, American biologists William Dreyer and J. Claude Bennett suggested a novel hypothesis to explain antibody diversity. As they put it, this hypothesis was “radically different from anything found in modern molecular genetics.” They proposed that a single antibody is made by separate gene segments that are brought together by recombination. According to this model, many copies of each gene segment are present, each slightly different from the others. Only one copy of each gene segment ends up in the final, recombined antibody gene (and hence in the antibody protein). Diversity is therefore achieved by combining different copies of each segment in different B cells as they mature.
More than 10 years after Dreyer and Bennett proposed their model, direct experimental evidence showed that their hypothesis is correct (Fig. 43.13). As a B cell differentiates, different gene segments are joined in a process called genomic rearrangement that produces a specific antibody.
HOW DO WE KNOW?
FIG. 43.13
How is antibody diversity generated?
BACKGROUND Humans produce an estimated 10 billion different antibodies. Antibodies are proteins, encoded by genes. However, there are only about 25,000 genes in the human genome. How then can humans (and other vertebrates) produce such a diversity of antibodies from a limited set of genes? This was one of the central problems in immunology until the 1960s and 1970s.
HYPOTHESIS In 1965, American biologists William Dreyer and J. Claude Bennett proposed a model in which there are many copies of two gene segments, which they called the V (variable) and C (constant) segments. An antibody gene is assembled by recombining a single copy of each of these segments. At the time, there was no direct experimental evidence to support this hypothesis.
EXPERIMENT In 1976, Japanese immunologists Nobumichi Hozumi and Susumu Tonegawa tested the Dreyer and Bennett hypothesis. Using mice as a model system, they isolated DNA from embryonic cells (before recombination was thought to occur) and from adult B-
RESULTS Hozumi and Tonegawa found that the V and C regions were far apart in the DNA of embryonic cells and close together in adult cells, suggesting that the gene segments were brought together during B cell differentiation.
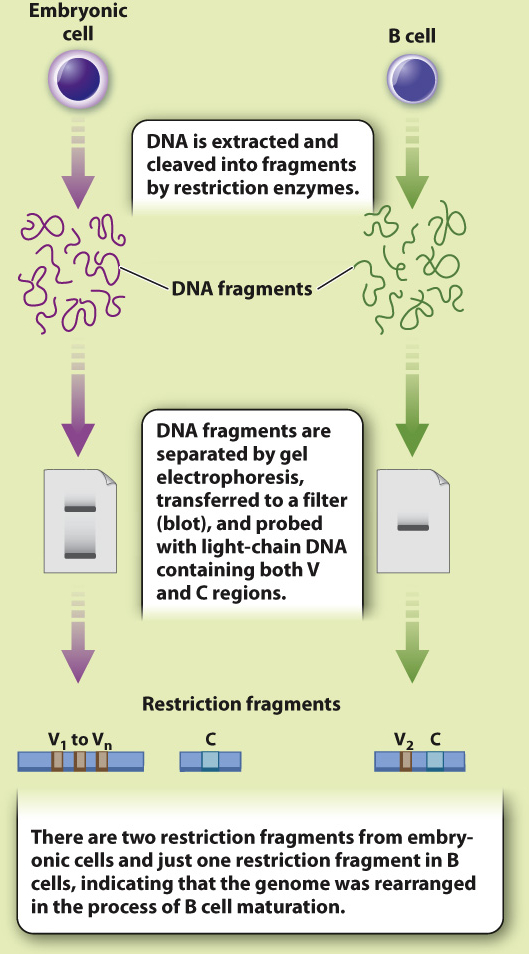
CONCLUSION Antibody genes are assembled by recombination of individual gene segments, providing a mechanism for generating antibody diversity.
FOLLOW-
SOURCES Dreyer, W. J., and J. C. Bennett. 1965. “The Molecular Basis of Antibody Formation: A Paradox.” Proceedings of the National Academy of Sciences 54:864–
Genes for H chains are composed of multiple different V (variable), D (diversity), J (joining), and C (constant) gene segments (Fig. 43.14). Genes for mice H chains, for example, can be composed of any of 100 different V segments, 30 D segments, 6 J segments, and 8 C segments. During B cell differentiation, the DNA for all of these gene segments undergoes recombination so that just one of each type of segment is present in the DNA, and the intervening DNA is deleted. The assembled VDJ segment encodes the variable portion, and the C segment encodes the constant region. L chain genes also contain multiple copies of gene segments, but they do not have a D gene segment, so the VJ segment encodes the variable portion and the C segment encodes the constant region. The result of this process is that each B cell encodes a single H chain and a single L chain, and each B cell expresses a unique antibody.
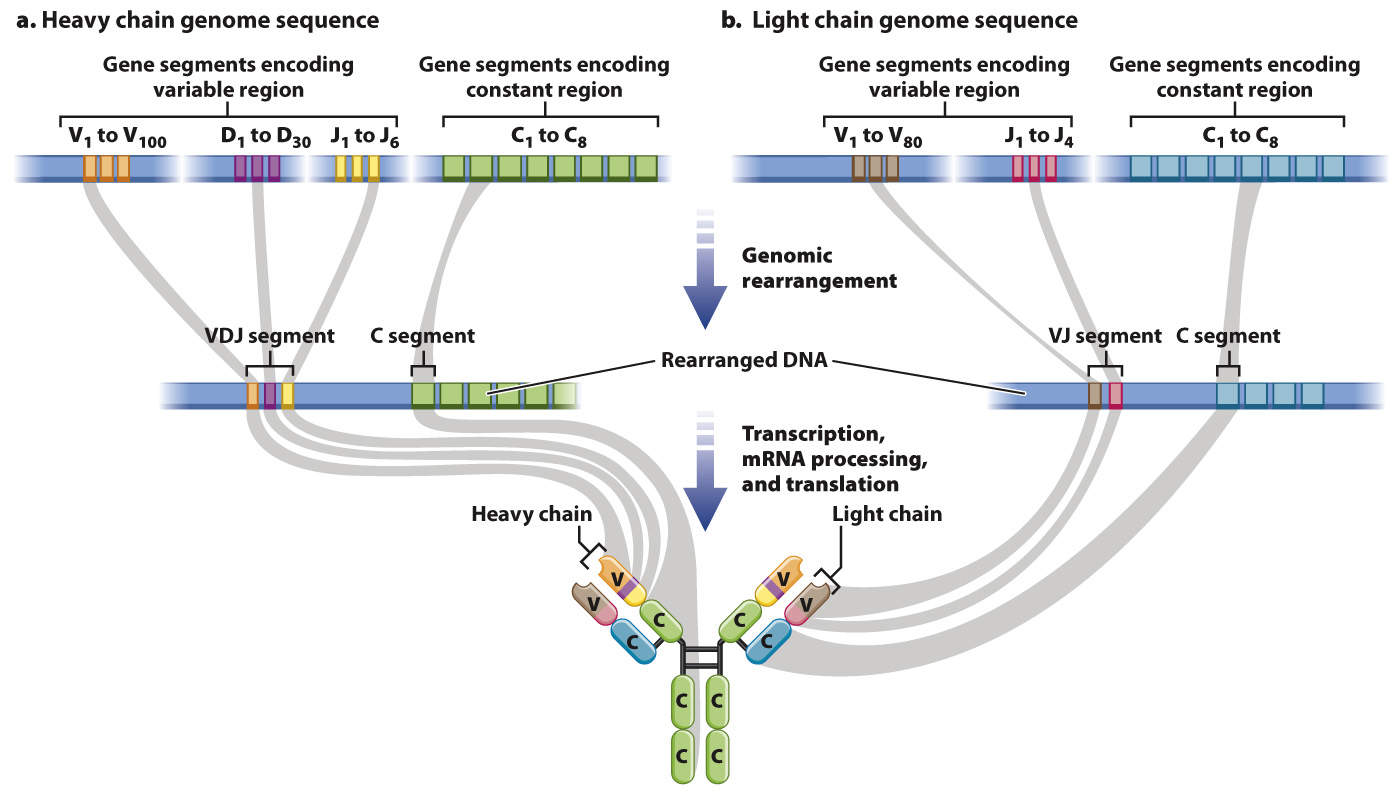
It is worth pausing here to consider the significance of this mechanism. We generally think of DNA as being a stable blueprint present in identical copies in all of the somatic cells in our bodies. B cells are an exception. As a result of genomic rearrangement, the DNA in each mature B cell is different from the DNA in every other mature B cell and different from the DNA in other cells in the body.
Genomic rearrangement is the primary mechanism that creates antibody diversity, but other mechanisms contribute as well. One is simply the association of different L and H chains to make a functional antibody. In addition, recombination is sometimes imprecise, creating different sequences at the junctions between the different gene segments. There are also mechanisms for adding or replacing nucleotides within VJ and VDJ segments. Finally, alternative splicing of mRNA (Chapter 3) allows a single B cell to express both membrane-
We have seen that B cells as a group produce an enormous diversity of antibodies, whereas a given B cell makes one and only one antibody. But B cells are diploid, containing maternal and paternal copies of the genes that encode for antibodies. Maternal and paternal alleles of the antibody genes are similar, but not identical. Therefore, a B cell able to use both alleles would make two different antibodies, yet B cells always make one. Remarkably, once one allele undergoes genomic rearrangement, the other allele is prevented from doing so. As a result, B cells function as if they have a single allele for each antibody gene—
The mechanisms we have considered pertain to humans and mice. Whereas B cells mature in the bone marrow in humans and mice, they mature in gut-
Quick Check 3 If a given B cell produces only one type of antibody, how do organisms produce a great diversity of antibodies?
Quick Check 3 Answer
Although a given B cell produces only one antibody, the entire population of B cells produces many different antibodies. Each B cell produces a different antibody as the result of a unique pattern of genomic rearrangement.