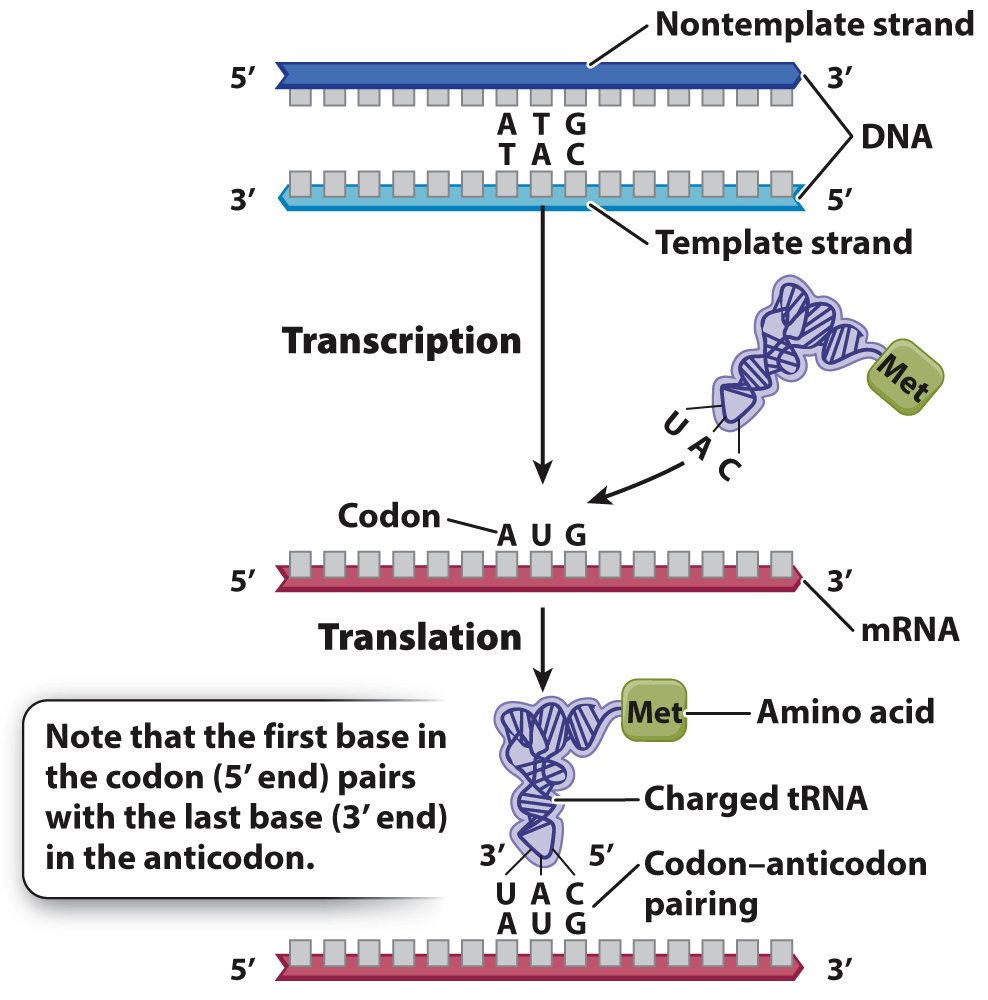
Translation uses many molecules found in all cells.
Translation requires many components. Well over 100 genes encode components needed for translation, some of which are shown in Fig. 4.11. What are these needed components? First, the cell needs ribosomes, which are complex structures of RNA and protein that bind with mRNA and are the site of translation. In prokaryotes, translation occurs as soon as the mRNA comes off the DNA template. In eukaryotes, the processes of transcription and translation are physically separated: Transcription takes place in the nucleus, and translation takes place in the cytoplasm.
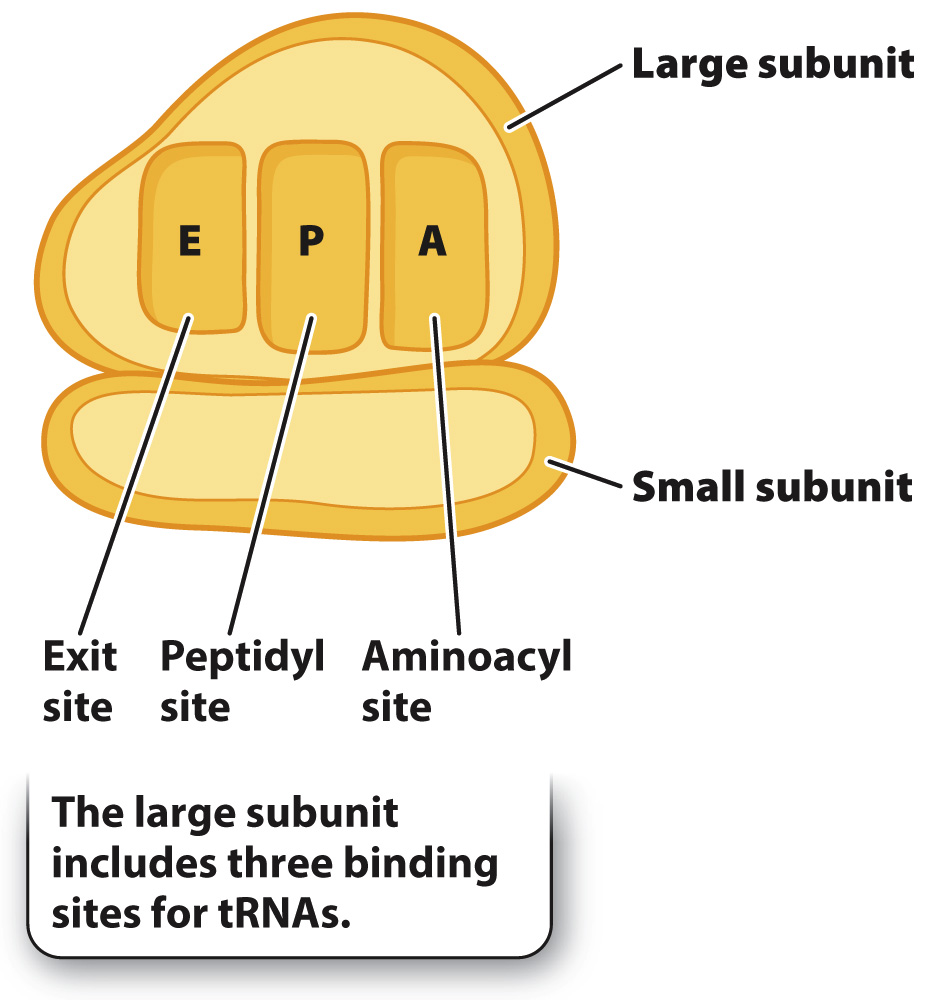
In both eukaryotes and prokaryotes, the ribosome consists of a small subunit and a large subunit, each composed of 1 to 3 types of ribosomal RNA and 20 to 50 types of ribosomal protein. Eukaryotic ribosomes are larger than prokaryotic ribosomes. As indicated in Fig. 4.12, the large subunit of the ribosome includes three binding sites for molecules of transfer RNA, which are called the A (aminoacyl) site, the P (peptidyl) site, and the E (exit) site.
A major role of the ribosome is to ensure that, when the mRNA is in place on the ribosome, the sequence in the mRNA coding for amino acids is read in successive, non-
THEBIGBOYSAWTHEBADMANRUN
Each non-
In the example above, it is clear that the sentence begins with THE. However, in a long linear mRNA molecule, the ribosome could begin at any nucleotide. As an analogy, if we knew that the letters THE were the start of the phrase, then we would know immediately how to read
ZWTHEBIGBOYSAWTHEBADMANRUN
However, without knowing where to start reading this string of letters, we could find three ways to break the sentence into three-
ZWT HEB IGB OYS AWT HEB ADM ANR UN
Z WTH EBI GBO YSA WTH EBA DMA NRU N
ZW THE BIG BOY SAW THE BAD MAN RUN
The different ways of parsing the string into three-
While the ribosome establishes the correct reading frame for the codons, the actual translation of each codon in the mRNA into one amino acid in the polypeptide is carried out by means of transfer RNA (tRNA). Transfer RNAs are small RNA molecules of 70 to 90 nucleotides (Fig. 4.13). Each has a characteristic self-
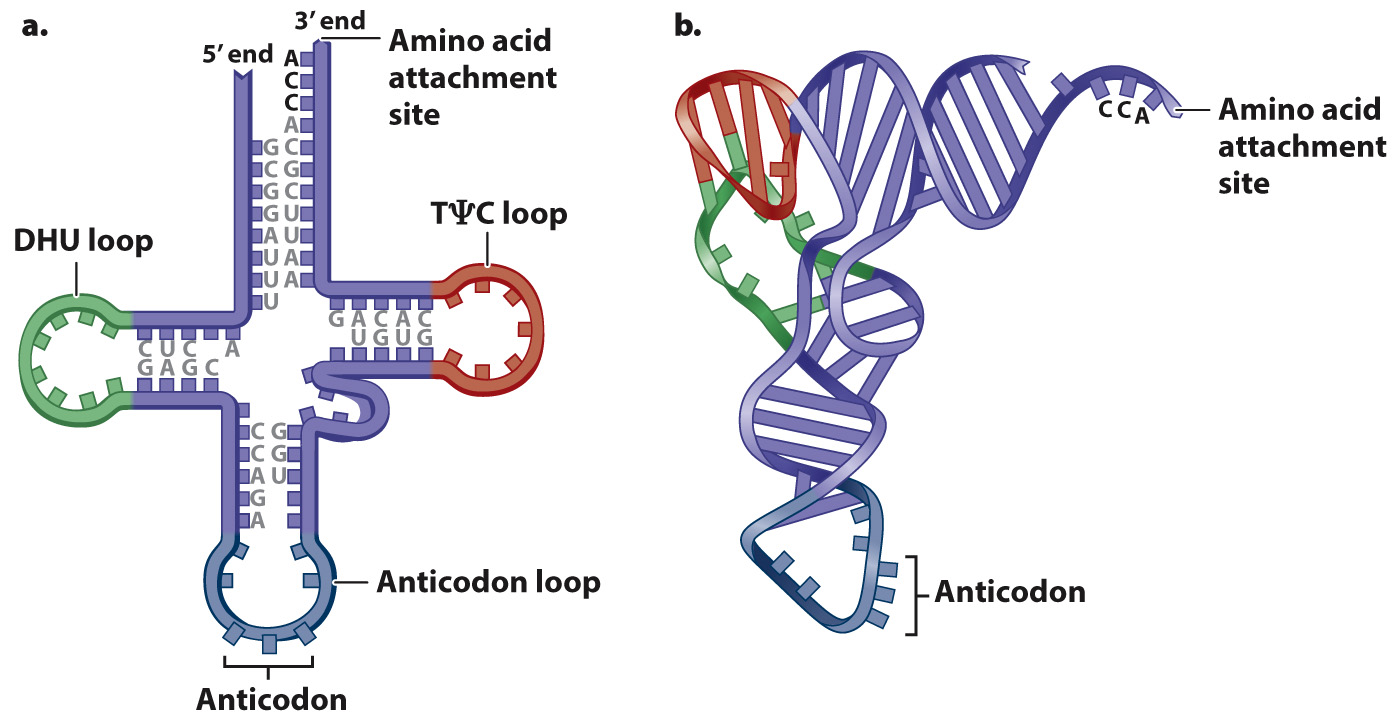
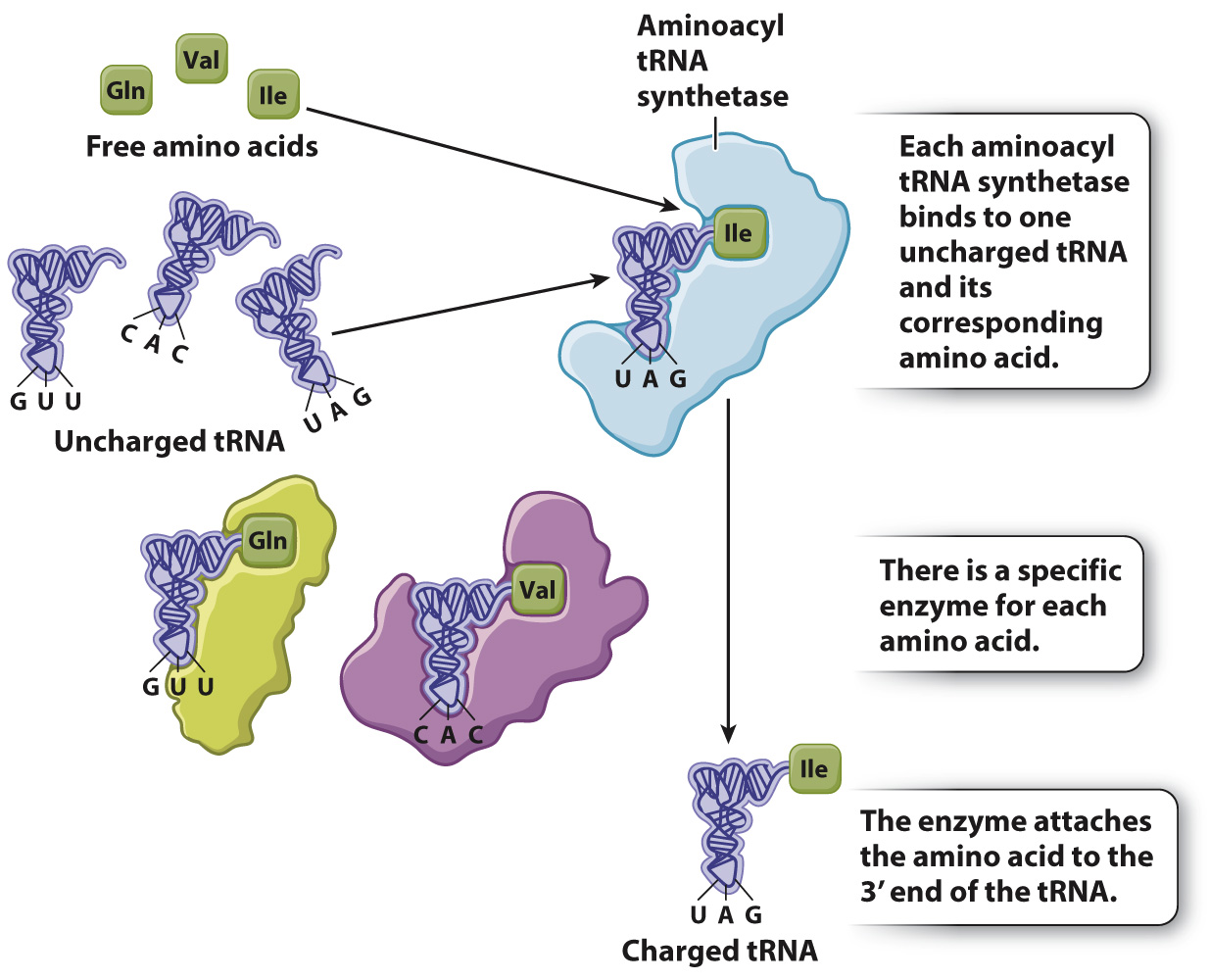
Each tRNA has the nucleotide sequence CCA at its 3′ end (Fig. 4.13), and the 3′ hydroxyl of the A is the attachment site for the amino acid corresponding to the anticodon. Enzymes called aminoacyl tRNA synthetases connect specific amino acids to specific tRNA molecules (Fig. 4.14). Therefore, these enzymes are directly responsible for actually translating the codon sequence in a nucleic acid to a specific amino acid in a polypeptide chain. Most organisms have one aminoacyl tRNA synthetase for each amino acid. The enzyme binds to multiple sites on any tRNA that has an anticodon corresponding to the amino acid, and it catalyzes formation of the covalent bond between the amino acid and tRNA. A tRNA that has no amino acid attached is said to be uncharged, and one with its amino acid attached is said to be charged. Amino acid tRNA synthetases are very accurate and attach the wrong amino acid far less often than 1 time in 10,000.
Although the specificity for attaching an amino acid to the correct tRNA is a property of aminoacyl tRNA synthetase, the specificity of DNA–