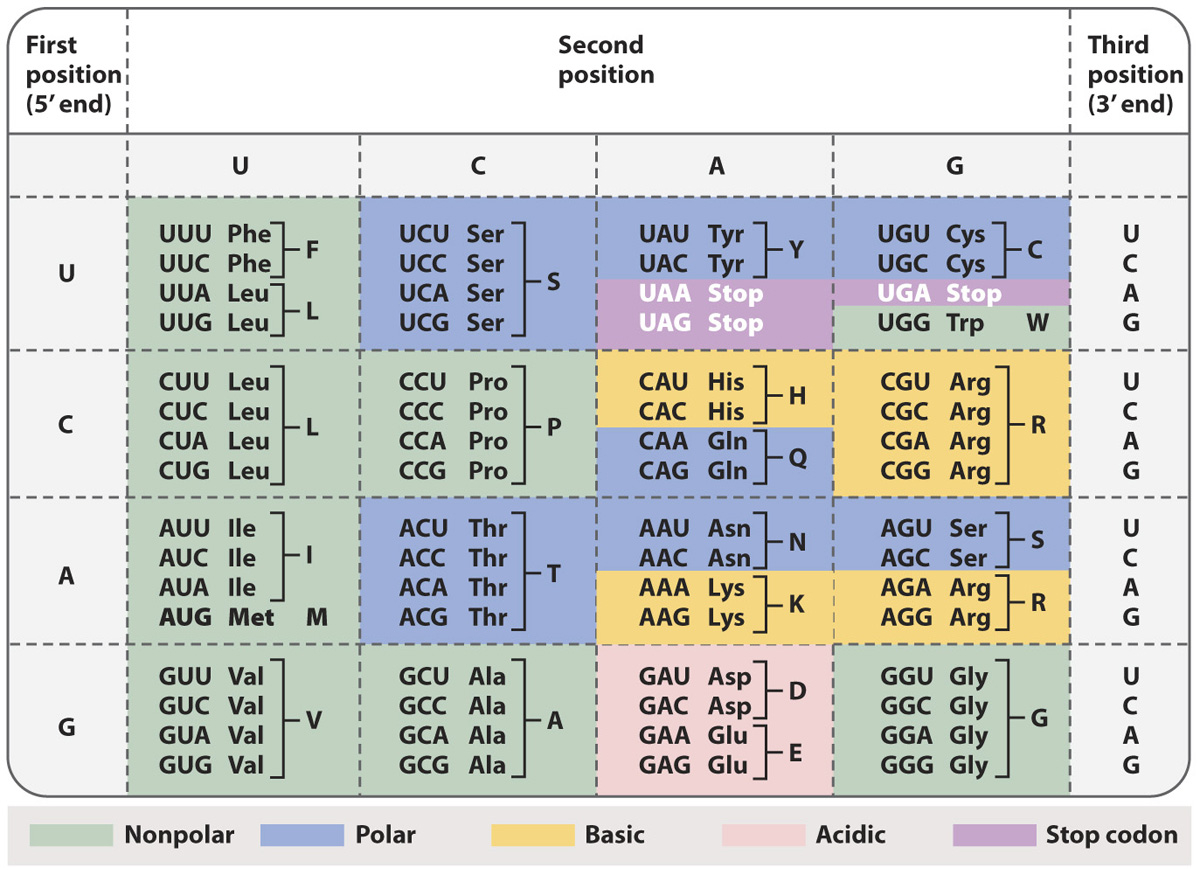
The genetic code shows the correspondence between codons and amino acids.
Fig. 4.15 shows how the codon AUG specifies the amino acid methionine (Met) by base pairing with the anticodon of a charged tRNA, denoted tRNAMet. Most codons specify an amino acid according to a genetic code. This code is sometimes called the “standard” genetic code because, while it is used by almost all cells, some minor differences are found in a few organisms as well as in mitochondria.
The codon at which translation begins is called the initiation codon, and it is coded by AUG, which specifies Met. The polypeptide is synthesized from the amino end to the carboxyl end, and so Met forms the amino end of any polypeptide being synthesized; however, in many cases the Met is cleaved off by an enzyme after synthesis is complete. The AUG codon is also used to specify the incorporation of Met at internal sites within the polypeptide chain.
As is apparent in Fig. 4.15, the AUG codon that initiated translation is preceded by a region in the mRNA that is not translated. The position of the initiator AUG codon in the mRNA establishes the reading frame that determines how the downstream codons (those following the AUG) are to be read.
Once the initial Met creates the amino end of a new polypeptide chain, the downstream codons are read one by one in non-
The standard genetic code was deciphered in the 1960s by a combination of techniques, but among the most ingenious were chemical methods for making synthetic RNAs of known sequence by American biochemist Har Gobind Khorana and his colleagues. This experiment is illustrated in Fig. 4.16.
HOW DO WE KNOW?
FIG. 4.16
How was the genetic code deciphered?
BACKGROUND The genetic code is the correspondence between three-
METHOD Khorana and his group made RNAs of known sequence. They then added these synthetic RNAs to a solution containing all of the other components needed for translation. By adjusting the concentration of magnesium and other factors, the researchers could get the ribosome to initiate synthesis with any codon, even if not AUG.
EXPERIMENT 1 AND RESULTS When a synthetic poly(U) was used as the mRNA, the resulting polypeptide was polyphenylalanine (Phe–
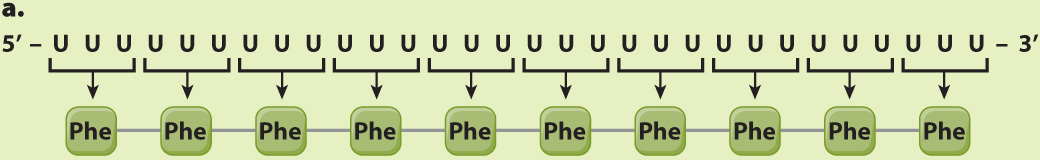
CONCLUSION The codon UUU corresponds to Phe. The poly(U) mRNA can be translated in three possible reading frames, depending on which U is the 5′ end of the start codon, but in each of them, all the codons are UUU.
EXPERIMENT 2 AND RESULTS When a synthetic mRNA with alternating U and C was used, the resulting polypeptide had alternating serine (Ser) and leucine (Leu):
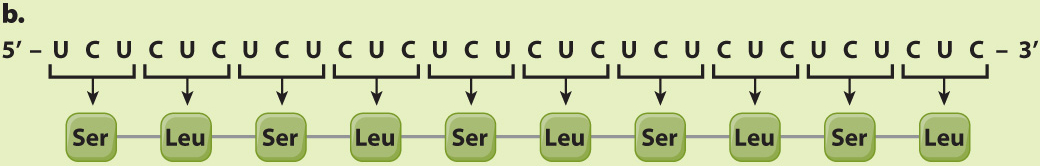
CONCLUSION Here again there are three reading frames, but each of them has alternating UCU and CUC codons. The researchers could not deduce from this result whether UCU corresponds to Ser and CUC to Leu or the other way around; the correct assignment came from experiments using other synthetic mRNA molecules.
EXPERIMENT 3 AND RESULTS When a synthetic mRNA with repeating UCA was used, three different polypeptides were produced—
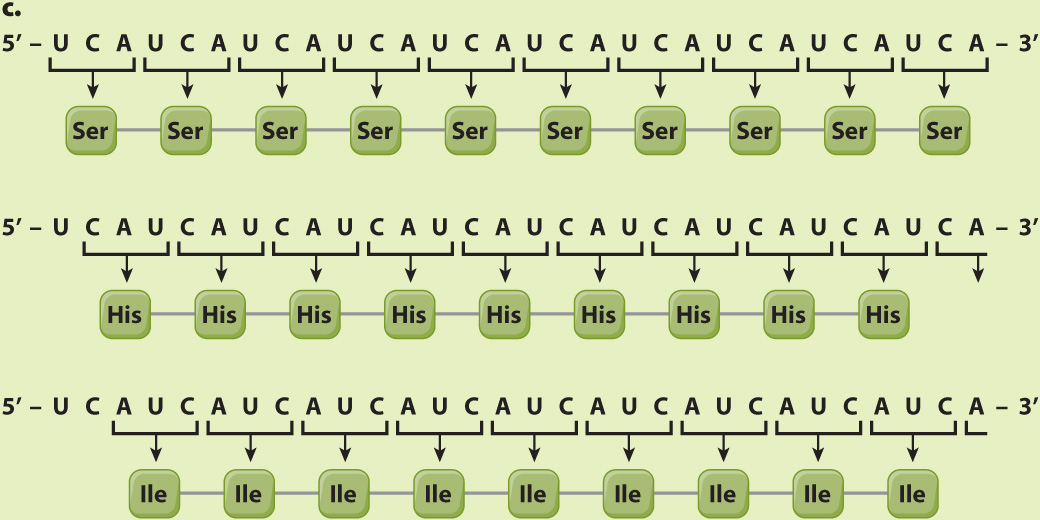
CONCLUSION The results do not reveal which of the three reading frames corresponds to which amino acid, but this was sorted out by studies of other synthetic polymers.
SOURCE Khorana, H. G. 1972. “Nucleic Acid Synthesis in the Study of the Genetic Code.” In Nobel Lectures, Physiology or Medicine 1963–
The standard genetic code shown in Table 4.1 has 20 amino acids specified by 64 codons. Many amino acids are therefore specified by more than one codon, and hence the genetic code is redundant, or degenerate. The redundancy has strong patterns, however:
The redundancy results almost exclusively from the third codon position.
When an amino acid is specified by two codons, they differ either in whether the third position is a U or a C (both pyrimidine bases), or in whether the third position is an A or a G (both purine bases).
When an amino acid is specified by four codons, the identity of the third codon position does not matter; it could be U, C, A, or G.
The chemical basis of these patterns results from two features of translation. First, in many tRNA anticodons the 5′ base that pairs with the 3′ (third) base in the codon is chemically modified into a form that can pair with two or more bases at the third position in the codon. Second, in the ribosome, there is less than perfect alignment between the third position of the codon and the base that pairs with it in the anticodon, so the requirements for base pairing are somewhat relaxed; this feature of the codon–
Quick Check 2 What polypeptide sequences would you expect to result from a synthetic mRNA with the repeating sequence 5′-UUUGGGUUUGGGUUUGGG-
Quick Check 2 Answer
The three reading frames are:
- UUU GGG UUU GGG…, which codes for repeating Phe–
Gly– Phe– Gly… - UUG GGU UUG GGU…, which codes for repeating Leu–
Gly– Leu– Gly… - UGG GUU UGG GUU…, which codes for repeating Trp–
Val– Trp– Val…