Translation consists of initiation, elongation, and termination.
Translation is usually divided into three separate processes. The first is initiation, in which the initiator AUG codon is recognized and Met is established as the first amino acid in the new polypeptide chain. The second process is elongation, in which successive amino acids are added one by one to the growing chain. And the third process is termination, in which the addition of amino acids stops and the completed polypeptide chain is released from the ribosome.
Initiation of translation (Fig. 4.17) requires a number of protein initiation factors that bind to the mRNA. In eukaryotes, one group of initiation factors binds to the 5′ cap that is added to the mRNA during processing. These recruit a small subunit of the ribosome, and other initiation factors bring up a transfer RNA charged with Met (Fig. 4.17a). The initiation complex then moves along the mRNA until it encounters the first AUG triplet. The position of this AUG establishes the translational reading frame.
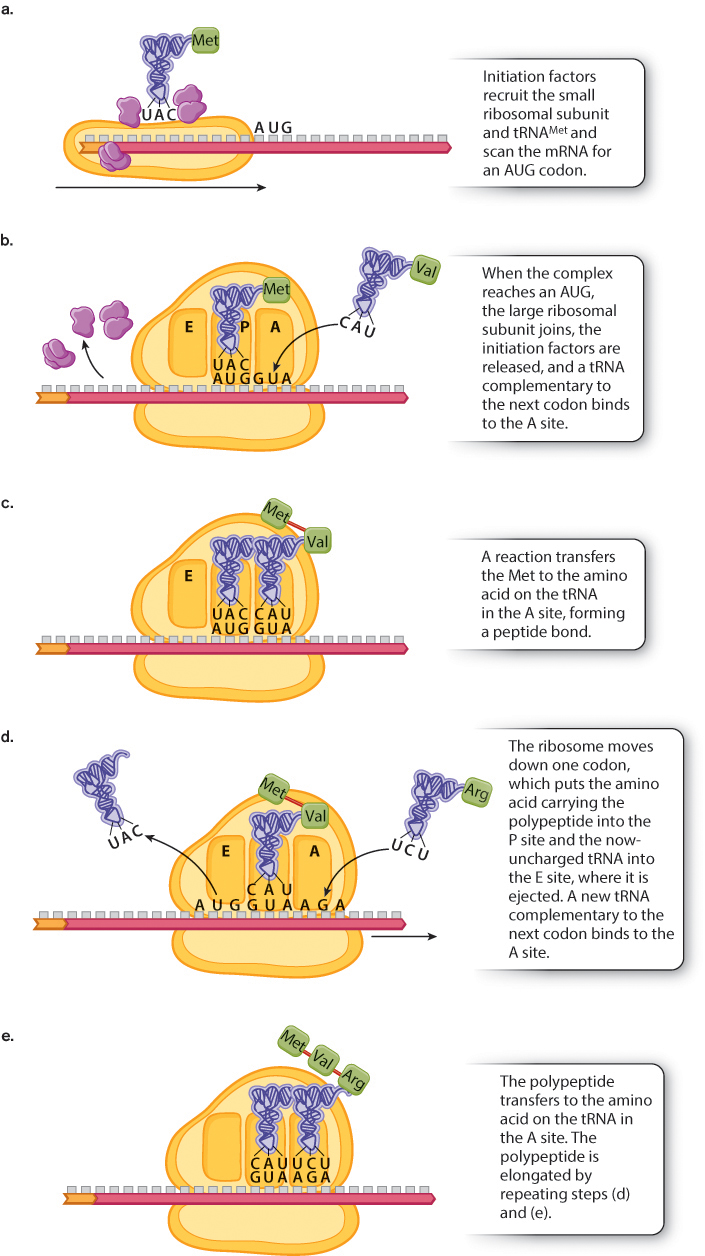
When the first AUG codon is encountered, a large ribosomal subunit joins the complex, the initiation factors are released, and the next tRNA is ready to join the ribosome (Fig. 4.17b). Note in Fig. 4.17b that the tRNAMet binds with the P (peptidyl) site in the ribosome and that the next tRNA in line comes in at the A (aminoacyl) site. Once the new tRNA is in place, a reaction takes place in which the bond connecting the Met to its tRNA is transferred to the amino group of the next amino acid in line, forming a peptide bond between the two amino acids (Fig. 4.17c). An RNA in the large subunit is the actual catalyst for this reaction. The new polypeptide is now attached to the tRNA in the A site. The ribosome then shifts one codon to the right (Fig. 4.17d). With this move, the uncharged tRNAMet shifts to the E site and is released into the cytoplasm, and the peptide-
At this point, steps (d) and (e) in Fig. 4.17 are repeated over and over again in the process of elongation, during which the polypeptide chain grows in length by the addition of successive amino acids. Ribosome movement along the mRNA and formation of the peptide bonds require energy, which is obtained with the help of proteins called elongation factors. Elongation factors are bound to GTP molecules, and break their high-
The elongation process shown in Fig. 4.17 continues until one of the stop codons (UAA, UAG, or UGA) is encountered; these codons signal termination of polypeptide synthesis. Termination takes place because the stop codons do not have corresponding tRNA molecules. Rather, when a stop codon is encountered, a protein release factor binds to the A site of the ribosome. The release factor causes the bond connecting the polypeptide to the tRNA to break, creating the carboxyl terminus of the polypeptide and completing the chain. Once the finished polypeptide is released, the small and large ribosomal subunits disassociate from the mRNA and from each other.
Although elongation and termination are very similar in prokaryotes and in eukaryotes, translation initiation differs between the two (Fig. 4.18). In eukaryotes, the initiation complex forms at the 5′ cap and scans along the mRNA until the first AUG is encountered (Fig. 4.18a). In prokaryotes, the mRNA molecules have no 5′ cap. Instead, the initiation complex is formed at one or more internal sequences present in the mRNA known as a Shine–
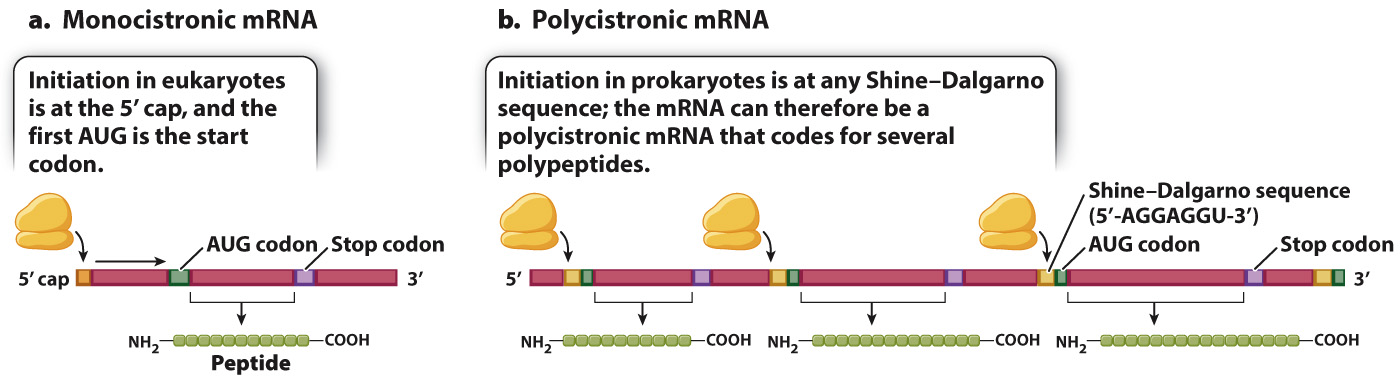
A polycistronic mRNA results from transcription of a group of functionally related genes located in tandem along the DNA and transcribed as a single unit from one promoter. This type of gene organization is known as an operon. Prokaryotes have many of their genes organized into operons because the production of a polycistronic mRNA allows all the protein products to be expressed together whenever they are needed. Typically, the genes organized into operons are those whose products are needed either for successive steps in the synthesis of an essential small molecule, such as an amino acid, or else for successive steps in the breakdown of a source of energy, such as a complex carbohydrate.
Quick Check 3 Bacterial DNA containing an operon encoding three enzymes is introduced into chromosomal DNA in yeast (a eukaryote) in such a way that it is properly flanked by a promoter and a transcriptional terminator. The bacterial DNA is transcribed and the RNA correctly processed, but only the protein nearest the promoter is produced. Can you suggest why?
Quick Check 3 Answer
With proper eukaryotic processing, the RNA transcript from the bacterial DNA will be capped at the 5′ end. The initiation complex will form at the 5′ cap and move along the mRNA until the first AUG codon is encountered, and then translation begins. When one of the termination codons is encountered, the polypeptide is released. Translation of the downstream polypeptides cannot take place because the Shine–