10.2 All Genetic Information Is Encoded in the Structure of DNA or RNA
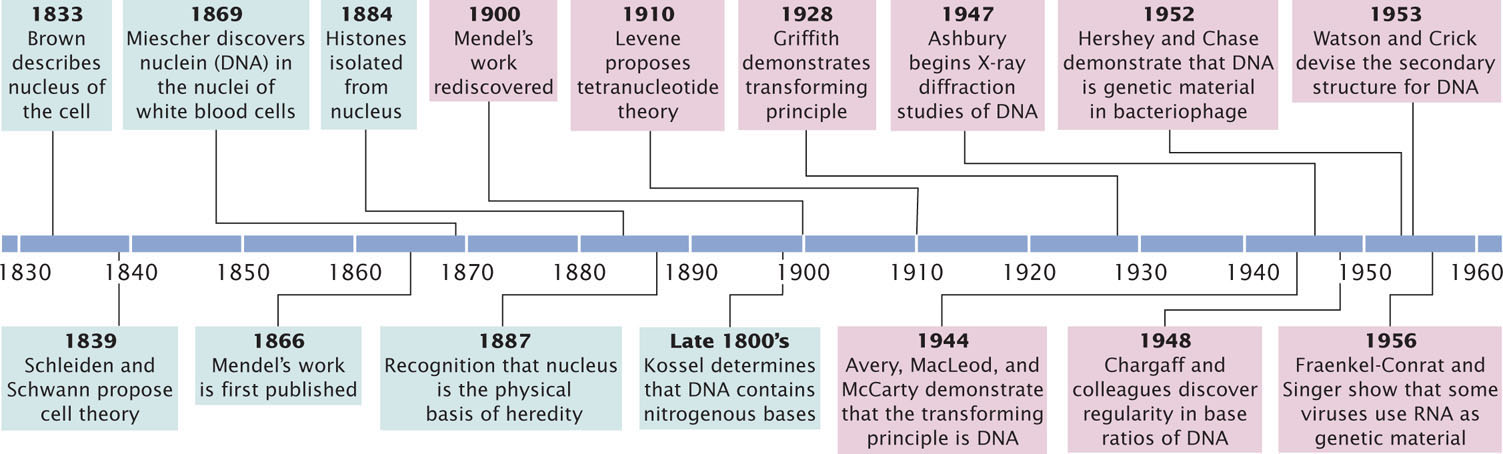
Although our understanding of how DNA encodes genetic information is relatively recent, the study of DNA structure stretches back more than 100 years (Figure 10.1).
Early Studies of DNA
In 1868, Johann Friedrich Miescher graduated from medical school in Switzerland. Influenced by an uncle who believed that the key to understanding disease lay in the chemistry of tissues, Miescher traveled to Tubingen, Germany, to study under Ernst Felix Hoppe-Seyler, an early leader in the emerging field of biochemistry. Under Hoppe-Seyler’s direction, Miescher turned his attention to the chemistry of pus, a substance of clear medical importance. Pus contains white blood cells with large nuclei; Miescher developed a method for isolating these nuclei. The minute amounts of nuclear material that he obtained were insufficient for a thorough chemical analysis, but he did establish that the nuclear material contained a novel substance that was slightly acidic and high in phosphorus. This material, which consisted of DNA and protein, Miescher called nuclein. The substance was later renamed nucleic acid by one of his students.
By 1887, several researchers independently concluded that the physical basis of heredity lies in the nucleus. Chromatin was shown to consist of nucleic acid and proteins, but which of these substances was actually the genetic information was not clear. In the late 1800s, further work on the chemistry of DNA was carried out by Albrecht Kossel, who determined that DNA contains four nitrogenous bases: adenine, cytosine, guanine, and thymine (abbreviated A, C, G, and T).
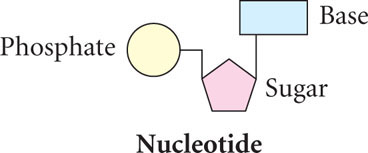
In the early twentieth century, the Rockefeller Institute in New York City became a center for nucleic acid research. Phoebus Aaron Levene joined the Institute in 1905 and spent the next 40 years studying the chemistry of DNA. He discovered that DNA consists of a large number of linked, repeating units, called nucleotides; each nucleotide contains a sugar, a phosphate, and a base.
Levene incorrectly proposed that DNA consists of a series of four-nucleotide units, each unit containing all four bases—adenine, guanine, cytosine, and thymine—in a fixed sequence. This concept, known as the tetranucleotide hypothesis, implied that the structure of DNA is not variable enough to be the genetic material. The tetranucleotide hypothesis contributed to the idea that protein is the genetic material because, with its 20 different amino acids, protein structure could be highly variable.
As additional studies of the chemistry of DNA were completed in the 1940s and 1950s, this notion of DNA as a simple, invariant molecule began to change. Erwin Chargaff and his colleagues carefully measured the amounts of the four bases in DNA from a variety of organisms and found that DNA from different organisms varies greatly in base composition. This finding disproved the tetranucleotide hypothesis. They discovered that, within each species, there is some regularity in the ratios of the bases: the amount of adenine is always equal to the amount of thymine (A = T), and the amount of guanine is always equal to the amount of cytosine (G = C; Table 10.1). These findings became known as Chargaff’s rules.
| Ratio | |||||||
| Source of DNA | A | T | G | C | A/T | G/C | (A + G)/(T + C) |
| E. coli | 26.0 | 23.9 | 24.9 | 25.2 | 1.09 | 0.99 | 1.04 |
| Yeast | 31.3 | 32.9 | 18.7 | 17.1 | 0.95 | 1.09 | 1.00 |
| Sea urchin | 32.8 | 32.1 | 17.7 | 18.4 | 1.02 | 0.96 | 1.00 |
| Rat | 28.6 | 28.4 | 21.4 | 21.5 | 1.01 | 1.00 | 1.00 |
| Human | 30.3 | 30.3 | 19.5 | 19.9 | 1.00 | 0.98 | 0.99 |
| *Percent in moles of nitrogeneous constituents per 100 g-atoms of phosphate in hydrolysate corrected for 100% recovery. From E. Chargaff and J. Davidson (eds). The Nucleic Acids, Vol 1. (New York: Academic press, 1955) | |||||||
CONCEPTS
Details of the structure of DNA were worked out by a number of scientists. At first, DNA was interpreted as being too regular in structure to carry genetic information but, by the 1940s, DNA from different organisms was shown to vary in its base composition.
 CONCEPT CHECK 2
CONCEPT CHECK 2Levene made which contribution to our understanding of DNA structure?
- Determined that the nucleus contains DNA.
- He determined that DNA contains four nitrogenous bases.
- He determined that DNA consists of nucleotides.
- He determined that the nucleotide bases of DNA are present in regular ratios.
DNA As the Source of Genetic Information
While chemists were working out the structure of DNA, biologists were attempting to identify the source of genetic information. Mendel identified the basic rules of heredity in 1866, but he had no idea about the physical nature of hereditary information. By the early 1900s, biologists had concluded that genes resided on chromosomes, which were known to contain both DNA and protein. Two sets of experiments, one conducted on bacteria and the other on viruses, provided pivotal evidence that DNA, rather than protein, was the genetic material.
The Discovery of the Transforming Principle
The first clue that DNA was the carrier of hereditary information came with the demonstration that DNA was responsible for a phenomenon called transformation. This phenomenon was first observed in 1928 by Fred Griffith, an English physician whose special interest was the bacterium that causes pneumonia: Streptococcus pneumoniae. Griffith had succeeded in isolating several different strains of S. pneumoniae (type I, II, III, and so forth). In the virulent (disease-causing) forms of a strain, each bacterium is surrounded by a polysaccharide coat, which makes the bacterial colony appear smooth when grown on an agar plate; these forms are referred to as S, for smooth. Griffith found that these virulent forms occasionally mutated to nonvirulent forms, which lack a polysaccharide coat and produce a rough-appearing colony; these forms are referred to as R, for rough.
Griffith observed that small amounts of living type IIIS bacteria injected into mice caused the mice to develop pneumonia and die; when he examined the dead mice, he found large amounts of type IIIS bacteria in their blood (Figure 10.2a). When Griffith injected type IIR bacteria into mice, the mice lived, and no bacteria were recovered from their blood (Figure 10.2b). Griffith knew that boiling killed all the bacteria and destroyed their virulence; when he injected large amounts of heat-killed type IIIS bacteria into mice, the mice lived and no type IIIS bacteria were recovered from their blood (Figure 10.2c).
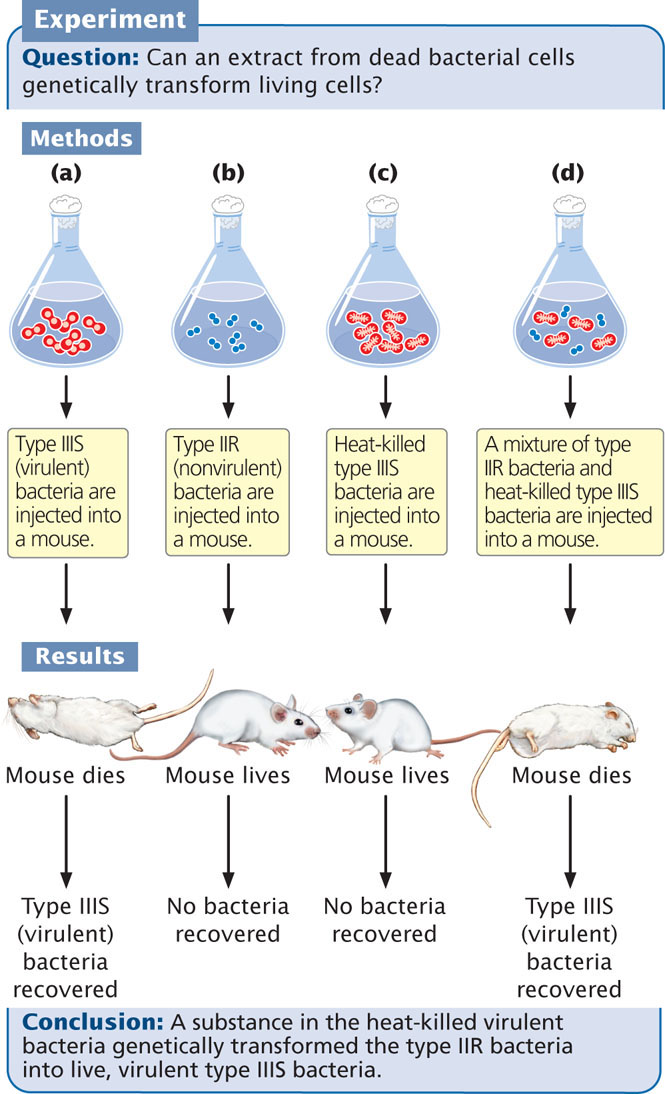
The results of these experiments were not unusual. However, Griffith got a surprise when he infected his mice with a small amount of living type IIR bacteria along with a large amount of heat-killed type IIIS bacteria. Because both the type IIR bacteria and the heat-killed type IIIS bacteria were nonvirulent, he expected these mice to live. Surprisingly, five days after the injections, the mice became infected with pneumonia and died (Figure 10.2d). When Griffith examined blood from the hearts of these mice, he observed live type IIIS bacteria. Furthermore, these bacteria retained their type IIIS characteristics through several generations; so the infectivity was heritable.
Griffith considered all of the possible interpretations of his results. First, it could have been the case that he had not sufficiently sterilized the type IIIS bacteria and thus a few live bacteria remained in the culture. Any live bacteria injected into the mice would have multiplied and caused pneumonia. Griffith knew that this possibility was unlikely, because he had used only heat-killed type IIIS bacteria in the control experiment, and they never produced pneumonia in the mice.
A second interpretation was that the live, type IIR bacteria had mutated to the virulent S form. Such a mutation would cause pneumonia in the mice, but it would produce type IIS bacteria, not the type IIIS that Griffith found in the dead mice. Because type II and type III bacteria differ in a number of traits, many mutations would be required for type II bacteria to mutate to type III bacteria, and the chance of all the mutations occurring simultaneously was impossibly low.
Griffith concluded that the type IIR bacteria had somehow been transformed, acquiring the genetic virulence of the dead type IIIS bacteria. This transformation had produced a permanent, genetic change in the bacteria. Although Griffith didn’t understand the nature of this transformation, he theorized that some substance in the polysaccharide coat of the dead bacteria might be responsible. He called this substance the transforming principle.  TRY PROBLEM 19
TRY PROBLEM 19
Identification of the Transforming Principle
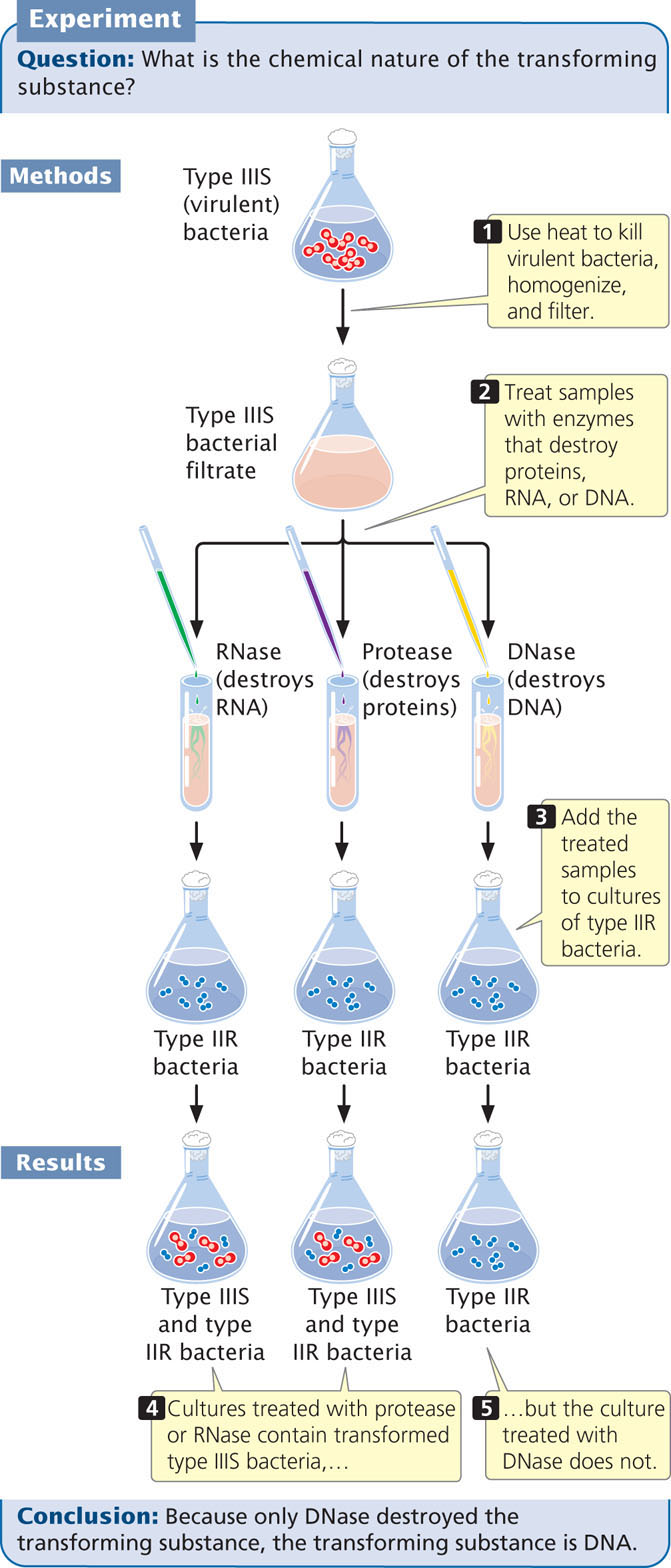
At the time of Griffith’s report, Oswald Avery (see Figure 10.1) was a microbiologist at the Rockefeller Institute. At first Avery was skeptical but, after other microbiologists successfully repeated Griffith’s experiments with other bacteria, Avery set out to identify the nature of the transforming substance.
After 10 years of research, Avery, Colin MacLeod, and Maclyn McCarty succeeded in isolating and partially purifying the transforming substance. They showed that it had a chemical composition closely matching that of DNA and quite different from that of proteins. Enzymes such as trypsin and chymotrypsin, known to break down proteins, had no effect on the transforming substance. Ribonuclease, an enzyme that destroys RNA, also had no effect. Enzymes capable of destroying DNA, however, eliminated the biological activity of the transforming substance (Figure 10.3). Avery, MacLeod, and McCarty showed that the transforming substance precipitated at about the same rate as purified DNA and that it absorbed ultraviolet light at the same wavelengths as DNA. These results, published in 1944, provided compelling evidence that the transforming principle—and therefore genetic information—resides in DNA. However, new theories in science are rarely accepted on the basis of a single experiment, and many biologists continued to prefer the hypothesis that the genetic material is protein.
CONCEPTS
The process of transformation indicates that some substance—the transforming principle—is capable of genetically altering bacteria. Avery, MacLeod, and McCarty demonstrated that the transforming principle is DNA, providing the first evidence that DNA is the genetic material.
 CONCEPT CHECK 3
CONCEPT CHECK 3If Avery, MacLeod, and McCarty had found that samples of heat-killed bacteria treated with RNase and DNase transformed bacteria, but samples treated with protease did not, what conclusion would they have made?
- Protease carries out transformation.
- RNA and DNA are the genetic materials.
- Protein is the genetic material.
- RNase and DNase are necessary for transformation.
The Hershey–Chase Experiment
A second piece of evidence that indicated DNA was the genetic material resulted from a study of the T2 virus conducted by Alfred Hershey and Martha Chase. The T2 virus is a bacteriophage (phage) that infects the bacterium Escherichia coli (Figure 10.4a). As stated in Chapter 9, a phage reproduces by attaching to the outer wall of a bacterial cell and injecting its DNA into the cell, where it replicates and directs the cell to synthesize phage protein. The phage DNA becomes encapsulated within the proteins, producing progeny phages that lyse (break open) the cell and escape (Figure 10.4b).
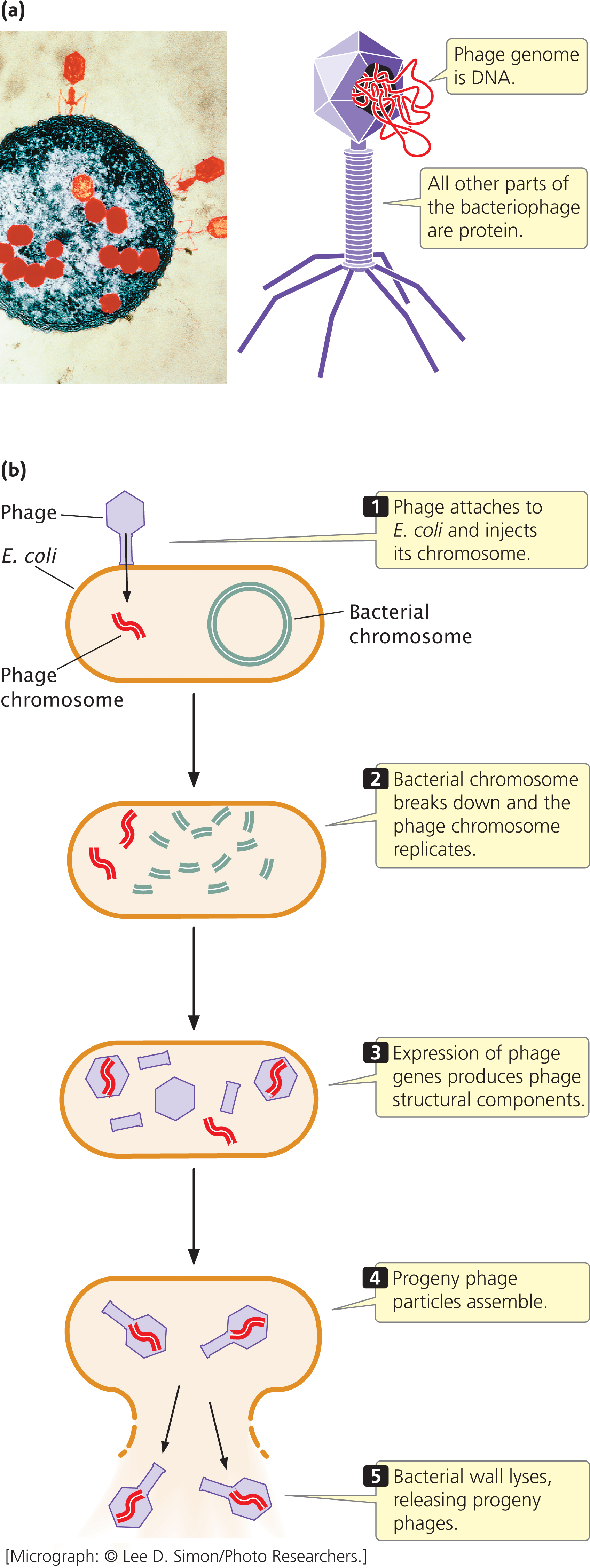
At the time of the Hershey–Chase study (their paper was published in 1952), biologists did not understand exactly how phages reproduce. What they did know was that the T2 phage is approximately 50% protein and 50% DNA, that a phage infects a cell by first attaching to the cell wall, and that progeny phages are ultimately produced within the cell. Because the progeny carry the same traits as the infecting phage, genetic material from the infecting phage must be transmitted to the progeny, but how this genetic transmission takes place was unknown.
Hershey and Chase designed a series of experiments to determine whether the phage protein or the phage DNA is transmitted in phage reproduction. To follow the fate of protein and DNA, they used radioactive forms, or isotopes, of phosphorus and sulfur. A radioactive isotope can be used as a tracer to identify the location of a specific molecule because any molecule containing the isotope will be radioactive and therefore easily detected. DNA contains phosphorus but not sulfur, so Hershey and Chase used 32P to follow phage DNA during reproduction. Protein contains sulfur but not phosphorus, so they used 35S to follow the protein.
Hershey and Chase grew one batch of E. coli in a medium containing 32P and infected the bacteria with T2 phage so that all the new phages would have DNA labeled with 32P (Figure 10.5). They grew a second batch of E. coli in a medium containing 35S and infected these bacteria with T2 phage so that all these new phages would have protein labeled with 35S. Hershey and Chase then infected separate batches of unlabeled E. coli with the 35S- and 32P-labeled phages. After allowing time for the phages to infect the cells, they placed the E. coli cells in a blender and sheared off the now-empty protein coats from the cell walls. They separated out the protein coats and cultured the infected bacterial cells.
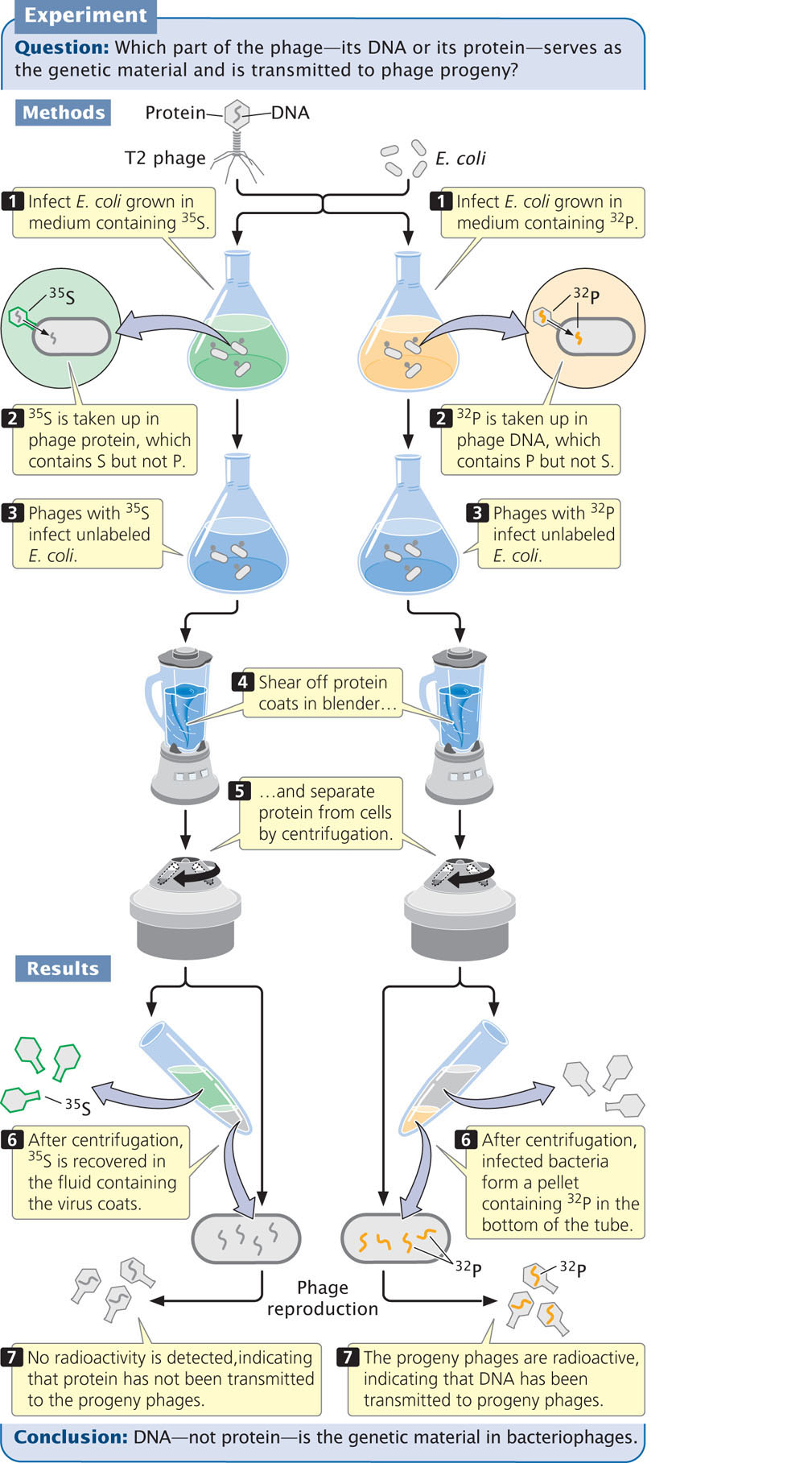
When phages labeled with 35S infected the bacteria, most of the radioactivity was detected in the protein coats and little was detected in the cells. Furthermore, when new phages emerged from the cell, they contained almost no 35S (see Figure 10.5). This result indicated that the protein component of a phage does not enter the cell and is not transmitted to progeny phages.
In contrast, when Hershey and Chase infected bacteria with 32P-labeled phages and removed the protein coats, the bacteria were radioactive. Most significantly, after the cells lysed and new progeny phages emerged, many of these phages emitted radioactivity from 32P, demonstrating that DNA from the infecting phages had been passed on to the progeny (see Figure 10.5). These results confirmed that DNA, not protein, is the genetic material of phages.  TRY PROBLEM 24
TRY PROBLEM 24
CONCEPTS
Using radioactive isotopes, Hershey and Chase traced the movement of DNA and protein during phage infection. They demonstrated that DNA, not protein, enters the bacterial cell during phage reproduction and that only DNA is passed on to progeny phages.
 CONCEPT CHECK 4
CONCEPT CHECK 4Could Hershey and Chase have used a radioactive isotope of carbon instead of 32P? Why or why not?
Watson and Crick′s Discovery of the Three-Dimensional Structure of DNA
The experiments on the nature of the genetic material set the stage for one of the most important advances in the history of biology—the discovery of the three-dimensional structure of DNA by James Watson and Francis Crick in 1953.
Before Watson and Crick’s breakthrough, much of the basic chemistry of DNA had already been determined by Miescher, Kossel, Levene, Chargaff, and others, who had established that DNA consists of nucleotides and that each nucleotide contains a sugar, a base, and a phosphate group. However, how the nucleotides fit together in the three-dimensional structure of the molecule was not at all clear.
In 1947, William Astbury began studying the three-dimensional structure of DNA by using a technique called X-ray diffraction (Figure 10.6), in which X-rays beamed at a molecule are reflected in specific patterns that reveal aspects of the structure of the molecule. However, his diffraction images did not provide enough resolution to reveal the structure. A research group at King’s College in London, led by Maurice Wilkins, also used X-ray diffraction to study DNA. Working in Wilkins’ laboratory, Rosalind Franklin obtained strikingly better images of the molecule. However, Wilkins and Franklin’s progress in developing a complete structure of the molecule was impeded by the personal discord that existed between them.
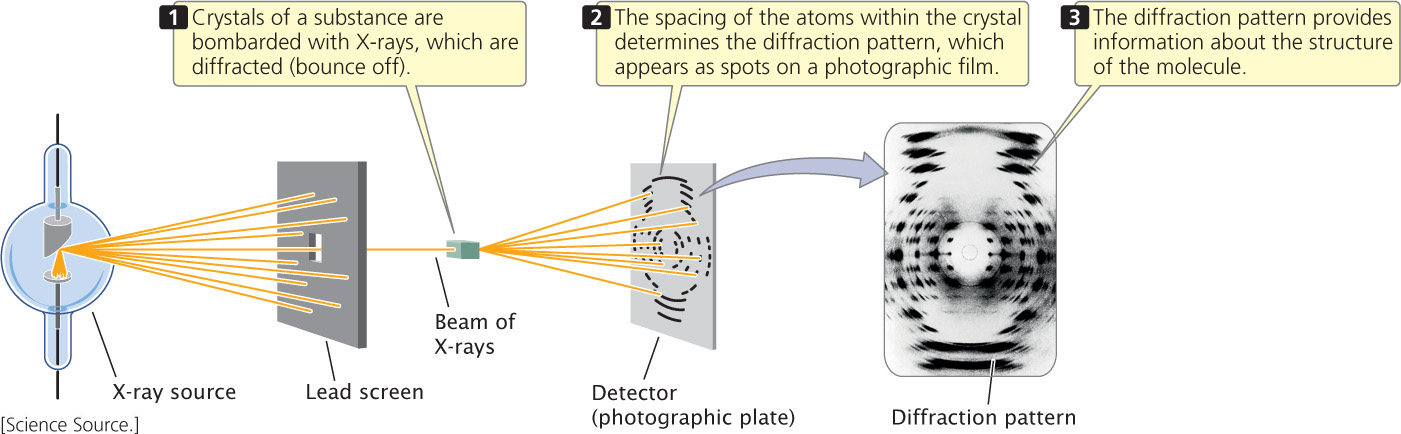
Watson and Crick investigated the structure of DNA, not by collecting new data but by using all available information about the chemistry of DNA to construct molecular models (Figure 10.7a). They used the excellent X-ray diffraction images taken by Rosalind Franklin (Figure 10.7b) and by applying the laws of structural chemistry they were able to limit the number of possible structures that DNA could assume. They tested various structures by building models made of wire and metal plates. With their models, they were able to see whether a structure was compatible with chemical principles and with the X-ray images.

The key to solving the structure came when Watson recognized that an adenine base could bond with a thymine base and that a guanine base could bond with a cytosine base; these pairings accounted for the base ratios that Chargaff had discovered earlier. The model developed by Watson and Crick showed that DNA consists of two strands of nucleotides that run in opposite directions (are antiparallel) and wind around each other to form a right-handed helix, with the sugars and phosphates on the outside and the bases in the interior. They recognized that the double-stranded structure of DNA with its specific base pairing provided an elegant means by which the DNA can be replicated. Watson and Crick published an electrifying description of their model in Nature in 1953. At the same time, Wilkins and Franklin each published their X-ray diffraction data, which demonstrated experimentally the hypothesis that DNA was helical in structure.
Many have called the solving of DNA’s structure the most important biological discovery of the twentieth century. For their discovery, Watson and Crick, along with Maurice Wilkins, were awarded a Nobel Prize in 1962. Rosalind Franklin had died of cancer in 1958 and thus could not be considered a candidate for the shared prize, but many scholars and historians believe that she should share equal credit for solving the structure of DNA.
After the discovery of DNA’s structure, much research focused on how genetic information is encoded within the base sequence and how this information is copied and expressed. Even today, the details of DNA structure and function continue to be the subject of active research.
CONCEPTS
By collecting existing information about the chemistry of DNA and building molecular models, Watson and Crick were able to discover the three-dimensional structure of the DNA molecule.
 CONCEPT CHECK 5
CONCEPT CHECK 5What did Watson and Crick use to help solve the structure of DNA?
- X-ray diffraction.
- Laws of structural chemistry
- Models of DNA
- All the above
RNA As Genetic Material
In most organisms, DNA carries the genetic information. However, a few viruses use RNA, not DNA, as their genetic material. This was demonstrated in 1956 by Heinz Fraenkel-Conrat and Bea Singer, who worked with the tobacco mosaic virus (TMV), a virus that infects and causes disease in tobacco plants (Figure 10.8). The tobacco mosaic virus possesses a single molecule of RNA surrounded by a helically arranged cylinder of protein molecules. Fraenkel-Conrat found that, after separating the RNA and protein of TMV, he could remix the RNA and protein of different strains of TMV and obtain intact, infectious viral particles.
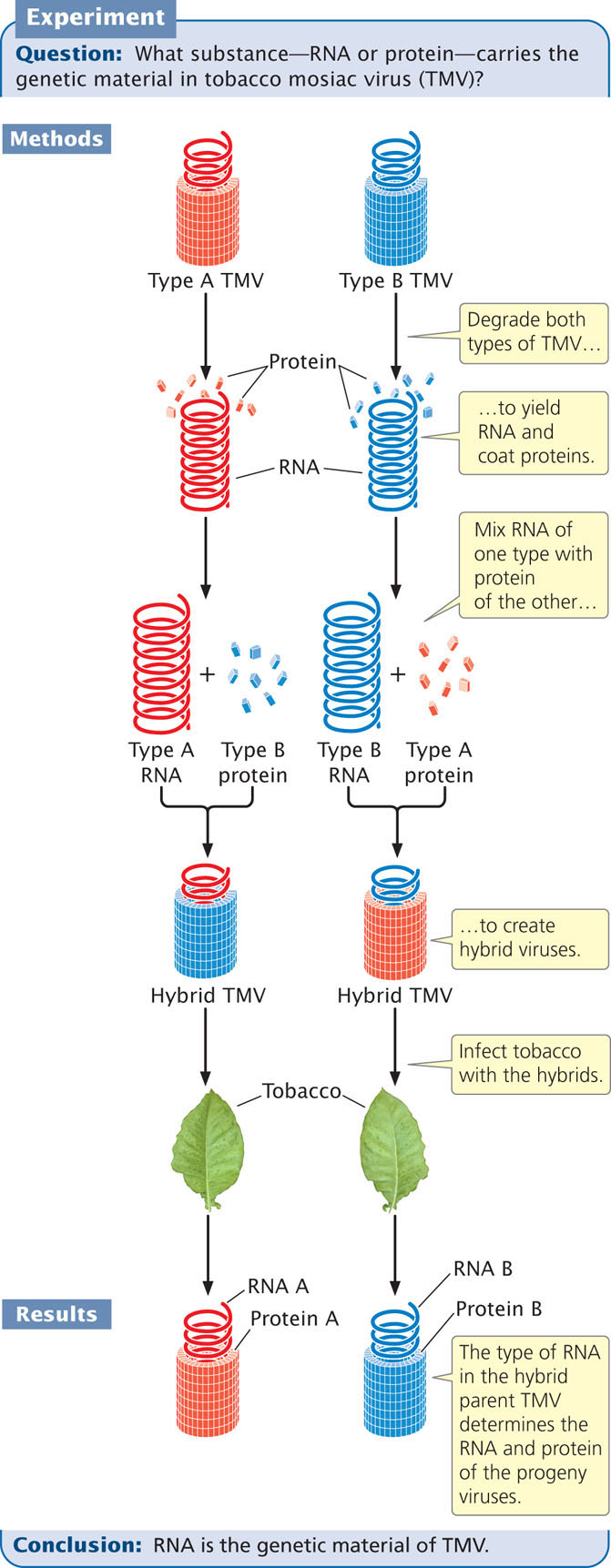
With Singer, Fraenkel-Conrat then created hybrid viruses by mixing RNA and protein from different strains of TMV. When these hybrid viruses infected tobacco leaves, new viral particles were produced. The new viral progeny were identical with the strain from which the RNA had been isolated and did not exhibit the characteristics of the strain that donated the protein. These results showed that RNA carries the genetic information in TMV.
Also in 1956, Alfred Gierer and Gerhard Schramm demonstrated that just RNA isolated from TMV is sufficient to infect tobacco plants and direct the production of new TMV particles. This finding confirmed that RNA carries genetic instructions.  TRY PROBLEM 18
TRY PROBLEM 18
CONCEPTS
RNA serves as the genetic material in some viruses.