Application Questions and Problems
Section 12.2
Question 12.20
Suppose a future scientist explores a distant planet and discovers a novel form of double-stranded nucleic acid. When this nucleic acid is exposed to DNA polymerases from E. coli, replication takes place continuously on both strands. What conclusion can you make about the structure of this novel nucleic acid?
Question 12.21
Phosphorus is required to synthesize the deoxyribonucleoside triphosphates used in DNA replication. A geneticist grows some E. coli in a medium containing nonradioactive phosphorous for many generations. A sample of the bacteria is then transferred to a medium that contains a radioactive isotope of phosphorus (32P). Samples of the bacteria are removed immediately after the transfer and after one and two rounds of replication. Assume that newly synthesized DNA contains 32P and the original DNA contains nonradioactive phosphorous. What will be the distribution of radioactivity in the DNA of the bacteria in each sample? Will radioactivity be detected in neither, one, or both strands of the DNA?
Question 12.22
A line of mouse cells is grown for many generations in a medium with 15N. Cells in G1 are then switched to a new medium that contains 14N. Draw a pair of homologous chromosomes from these cells at the following stages, showing the two strands of DNA molecules found in the chromosomes. Use different colors to represent strands with 14N and 15N.
- a. Cells in G1, before switching to medium with 14N
- b. Cells in G2, after switching to medium with 14N
- c. Cells in anaphase of mitosis, after switching to medium with 14N
- d. Cells in metaphase I of meiosis, after switching to medium with 14N
- e. Cells in anaphase II of meiosis, after switching to medium with 14N
Question 12.23
A circular molecule of DNA contains 1 million base pairs. If the rate of DNA synthesis at a replication fork is 100,000 nucleotides per minute, how much time will theta replication require to completely replicate the molecule, assuming that theta replication is bidirectional? How long will replication of this circular chromosome take by rolling-circle replication? Ignore replication of the displaced strand in rolling-circle replication.
Question 12.24
A bacterium synthesizes DNA at each replication fork at a rate of 1000 nucleotides per second. If this bacterium completely replicates its circular chromosome by theta replication in 30 minutes, how many base pairs of DNA will its chromosome contain?
Section 12.3
Question 12.25
The following diagram represents a DNA molecule that is undergoing replication. Draw in the strands of newly synthesized DNA and identify (a) the polarity of newly synthesized strands, (b) the leading and lagging strands, (c) Okazaki fragments, and (d) RNA primers.
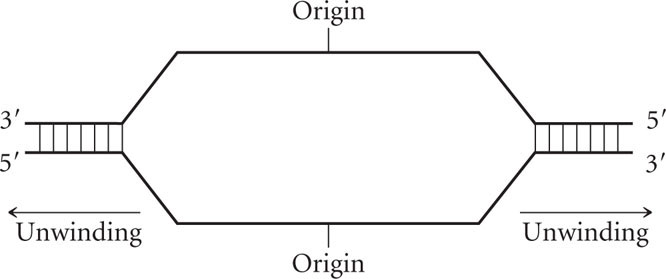
Question 12.26
In Figure 12.8, which is the leading strand and which is the lagging strand?
Question 12.27
What would be the effect on DNA replication of mutations that destroyed each of the following activities in DNA polymerase I?
- a. 3′→5′ exonuclease activity
- b. 5′→3′ exonuclease activity
- c. 5′→3′ polymerase activity
Question 12.28
Which of the DNA polymerases shown in Table 12.3 has the ability to proofread?
Question 12.29
How would DNA replication be affected in a bacterial cell that is lacking DNA gyrase?
Question 12.30
If the gene for primase were mutated so that no functional primase was produced, what would be the effect on theta replication? On rolling-circle replication?
Question 12.31
DNA polymerases are not able to prime replication, yet primase and other RNA polymerases can. Some geneticists have speculated that the inability of DNA polymerase to prime replication is due to its proofreading function. This hypothesis argues that proofreading is essential for the faithful transmission of genetic information and that, because DNA polymerases have evolved the ability to proofread, they cannot prime DNA synthesis. Explain why proofreading and priming functions in the same enzyme might be incompatible.
355
Section 12.4
Question 12.32
 Marina Melixetian and her colleagues suppressed the expression of Geminin protein in human cells by treating the cells with small interfering RNAs (siRNAs) complementary to Geminin messenger RNA (M. Melixetian et al. 2004. Journal of Cell Biology 165:473–482). (Small interfering RNAs form a complex with proteins and pair with complementary sequences on mRNAs; the complex then cleaves the mRNA, so there is no translation of the mRNA; pp. 403–404 in Chapter 14). Forty-eight hours after treatment with siRNA, the Geminin-depleted cells were enlarged and contained a single giant nucleus. Analysis of DNA content showed that many of these Geminin-depleted cells were 4n or greater. Explain these results.
Marina Melixetian and her colleagues suppressed the expression of Geminin protein in human cells by treating the cells with small interfering RNAs (siRNAs) complementary to Geminin messenger RNA (M. Melixetian et al. 2004. Journal of Cell Biology 165:473–482). (Small interfering RNAs form a complex with proteins and pair with complementary sequences on mRNAs; the complex then cleaves the mRNA, so there is no translation of the mRNA; pp. 403–404 in Chapter 14). Forty-eight hours after treatment with siRNA, the Geminin-depleted cells were enlarged and contained a single giant nucleus. Analysis of DNA content showed that many of these Geminin-depleted cells were 4n or greater. Explain these results.
Question 12.33
What results would be expected in the experiment outlined in Figure 12.17 if, during replication, all the original histone proteins remained on one strand of the DNA and new histones attached to the other strand?
Question 12.34
A number of scientists who study cancer treatment have become interested in telomerase. Why? How might cancer-drug therapies that target telomerase work?
Question 12.35
The enzyme telomerase is part protein and part RNA. What would be the most likely effect of a large deletion in the gene that encodes the RNA part of telomerase? How would the function of telomerase be affected?
Question 12.36
 Dyskeratosis congenita (DKC) is a rare genetic disorder characterized by abnormal fingernails and skin pigmentation, the formation of white patches on the tongue and cheek, and progressive failure of the bone marrow. An autosomal dominant form of DKC results from mutations in the gene that encodes the RNA component of telomerase. Tom Vulliamy and his colleagues examined 15 families with autosomal dominant DKC (T. Vulliamy et al. 2004. Nature Genetics 36:447–449). They observed that the median age of onset of DKC in parents was 37 years, whereas the median age of onset in the children of affected parents was 14.5 years. Thus, DKC in these families arose at progressively younger ages in successive generations, a phenomenon known as anticipation. The researchers measured telomere length of members of these families; the measurements are given in the adjoining table. Telomere length normally shortens with age, and so telomere length was adjusted for age. Note that the age-adjusted telomere length of all members of these families is negative, indicating that their telomeres are shorter than normal. For age-adjusted telomere length, the more negative the number, the shorter the telomere.
Dyskeratosis congenita (DKC) is a rare genetic disorder characterized by abnormal fingernails and skin pigmentation, the formation of white patches on the tongue and cheek, and progressive failure of the bone marrow. An autosomal dominant form of DKC results from mutations in the gene that encodes the RNA component of telomerase. Tom Vulliamy and his colleagues examined 15 families with autosomal dominant DKC (T. Vulliamy et al. 2004. Nature Genetics 36:447–449). They observed that the median age of onset of DKC in parents was 37 years, whereas the median age of onset in the children of affected parents was 14.5 years. Thus, DKC in these families arose at progressively younger ages in successive generations, a phenomenon known as anticipation. The researchers measured telomere length of members of these families; the measurements are given in the adjoining table. Telomere length normally shortens with age, and so telomere length was adjusted for age. Note that the age-adjusted telomere length of all members of these families is negative, indicating that their telomeres are shorter than normal. For age-adjusted telomere length, the more negative the number, the shorter the telomere.
| Parent telomere length | Child telomere length |
|---|---|
| –4.7 | –6.1 |
| –6.6 | |
| –6.0 | |
| –3.9 | –0.6 |
| –1.4 | –2.2 |
| –5.2 | –5.4 |
| –2.2 | –3.6 |
| –4.4 | –2.0 |
| –4.3 | –6.8 |
| –5.0 | –3.8 |
| –5.3 | –6.4 |
| –0.6 | –2.5 |
| –1.3 | –5.1 |
| –3.9 | |
| –4.2 | –5.9 |
- a. How does the telomere length of the parents compare with the telomere length of the children? (Hint: Calculate the average telomere length of all parents and the average telomere length of all children.)
- b. Explain why the telomeres of people with DKC are shorter than normal.
- c. Explain why DKC arises at an earlier age in subsequent generations.
Question 12.37
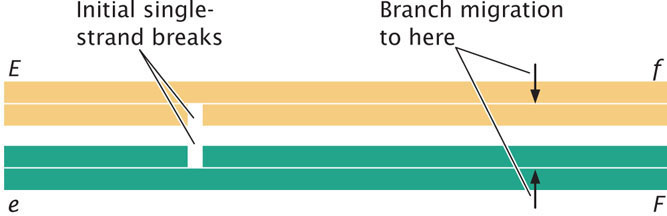
An individual is heterozygous at two loci (Ee Ff) and the genes are in repulsion. Assume that single-strand breaks and branch migration occur at the positions shown below. Using different colors to represent the two homologous chromosomes, draw out the noncrossover recombinant and crossover recombinant DNA molecules that will result from homologous recombination (Hint: see Figure 12.20).
356