Application Questions and Problems
Section 13.1
Question 13.14
An RNA molecule has the following percentages of bases: A = 23%, U = 42%, C = 21%, and G = 14%.
- a. Is this RNA single stranded or double stranded? How can you tell?
- b. What would be the percentages of bases in the template strand of the DNA that contains the gene for this RNA?
Section 13.2
Question 13.15
The following diagram represents DNA that is part of the RNA-coding sequence of a transcription unit. The bottom strand is the template strand. Give the sequence found on the RNA molecule transcribed from this DNA and identify the 5′ and 3′ ends of the RNA.
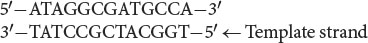
Question 13.16
For the RNA molecule shown in Figure 13.1a, write out the sequence of bases on the template and nontemplate strands of DNA from which this RNA is transcribed. Label the 5′ and 3′ ends of each strand.
Question 13.17
The following sequence of nucleotides is found in a single-stranded DNA template:
ATTGCCAGATCATCCCAATAGAT
Assume that RNA polymerase proceeds along this template from left to right.
- a. Which end of the DNA template is 5′ and which end is 3′?
- b. Give the sequence and identify the 5′ and 3′ ends of the RNA copied from this template.
Question 13.18
RNA polymerases carry out transcription at a much lower rate than that at which DNA polymerases carry out replication. Why is speed more important in replication than in transcription?
Question 13.19
Assume that a mutation occurs in the gene that codes for each of the following RNA polymerases. Match the mutation with possible effects by placing the correct letter in the blank below. There may be more than one effect for each mutated polymerase.
| A Mutation in the Gene That Codes for | Effects |
|---|---|
| RNA Polymerase I | ________ |
| RNA Polymerase II | ________ |
| RNA Polymerase III | ________ |
Possible Effects
- a. tRNA not synthesized
- b. some ribosomal RNA not synthesized
- c. ribosomal RNA not processed
- d. pre-mRNA not processed
- e. some mRNA molecules not degraded
- f. pre-mRNA not synthesized
Section 13.3
Question 13.20
Provide the consensus sequence for the first three actual sequences shown in Figure 13.10.
Question 13.21
Write the consensus sequence for the following set of nucleotide sequences.

Question 13.22
List at least five properties that DNA polymerases and RNA polymerases have in common. List at least three differences.
Question 13.23
Most RNA molecules have three phosphate groups at the 5′ end, but DNA molecules never do. Explain this difference.
Question 13.24
Write a hypothetical sequence of bases that might be found in the first 20 nucleotides of a promoter of a bacterial gene. Include both strands of DNA and identify the 5′ and 3′ ends of both strands. Be sure to include the start site for transcription and any consensus sequences found in the promoter.
Question 13.25
What would be the most likely effect of a mutation at the following locations in an E. coli gene?
- a. −8
- b. −35
- c. −20
- d. Start site
Question 13.26
A strain of bacteria possesses a temperature-sensitive mutation in the gene that encodes the sigma factor. The mutant bacteria produce a sigma factor that is unable to bind to RNA polymerase at elevated temperatures. What effect will this mutation have on the process of transcription when the bacteria are raised at elevated temperatures?
Question 13.27
On Figure 13.5, indicate the location of the promoters and terminators for genes a, b, and c.
Question 13.28
The following diagram represents a transcription unit on a DNA molecule.
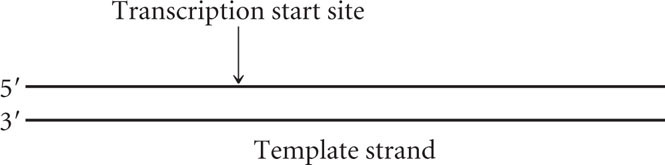
- a. Assume that this DNA molecule is from a bacterial cell. Draw the approximate location of the promoter and terminator for this transcription unit.
- b. Assume that this DNA molecule is from a eukaryotic cell. Draw the approximate location of an RNA polymerase II promoter.
Question 13.29
The following DNA nucleotides are found near the end of a bacterial transcription unit.
3′–AGCATACAGCAGACCGTTGGTCTGAAAAAAGCATACA–5′
- a. Mark the point at which transcription will terminate.
- b. Is this terminator rho independent or rho dependent?
- c. Draw a diagram of the RNA that will be transcribed from this DNA, including its nucleotide sequence and any secondary structures that form.
Question 13.30
A strain of bacteria possesses a temperature-sensitive mutation in the gene that encodes the rho subunit. At high temperatures, rho is not functional. When these bacteria are raised at elevated temperatures, which of the following effects would you expect to see? Explain your reasoning for accepting or rejecting each of these five options.
- a. Transcription does not take place.
- b. All RNA molecules are shorter than normal.
- c. All RNA molecules are longer than normal.
- d. Some RNA molecules are longer than normal.
- e. RNA is copied from both DNA strands.
Question 13.31
The following diagram represents one of the Christmas-tree-like structures as shown in Figure 13.3. On the diagram, identify parts a through i.
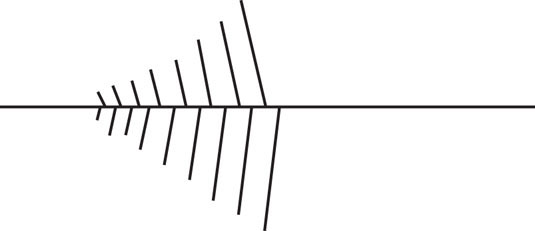
- a. DNA molecule
- b. 5′ and 3′ ends of the template strand of DNA
- c. At least one RNA molecule
- d. 5′ and 3′ ends of at least one RNA molecule
- e. Direction of movement of the transcription apparatus on the DNA molecule
- f. Approximate location of the promoter
- g. Possible location of a terminator
- h. Upstream and downstream directions
- i. Molecules of RNA polymerase (use dots to represent these molecules)
Question 13.32
Suppose that the string of A nucleotides following the inverted repeat in a rho-independent terminator was deleted but that the inverted repeat was left intact. How will this deletion affect termination? What will happen when RNA polymerase reached this region?
Section 13.4
Question 13.33
The following diagram represents a transcription unit in a hypothetical DNA molecule.
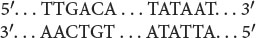
- a. On the basis of the information given, is this DNA from a bacterium or from a eukaryotic organism?
- b. If this DNA molecule is transcribed, which strand will be the template strand and which will be the nontemplate strand?
- c. Where, approximately, will the start site of transcription be?
Question 13.34
Computer programmers, working with molecular geneticists, have developed computer programs that can identify genes within long stretches of DNA sequences. Imagine that you are working with a computer programmer on such a project. On the basis of what you know about the process of transcription, what sequences should be used to identify the beginning and end of a gene with the use of this computer program?
Question 13.35
Through genetic engineering, a geneticist mutates the gene that encodes TBP in cultured human cells. This mutation destroys the ability of TBP to bind to the TATA box. Predict the effect of this mutation on cells that possess it.
Question 13.36
Elaborate repair mechanisms are associated with replication to prevent permanent mutations in DNA, yet no similar repair is associated with transcription. Can you think of a reason for this difference in replication and transcription? (Hint: Think about the relative effects of a permanent mutation in a DNA molecule compared with one in an RNA molecule.)