Answers to Concept Checks
- 1. b
- 2. The template strand is the DNA strand that is copied into an RNA molecule, whereas the nontemplate strand is not copied.
- 3. d
- 4. The sigma factor recognizes the promoter and controls the binding of RNA polymerase to the promoter.
- 5. a
- 6. Inverted repeats followed by a string of adenine nucleotides.
- 7. d
- 8. TFIID binds to the TATA box and helps to center the RNA polymerase over the start site of transcription.
- 9. Both processes use a protein that binds to the RNA molecule and moves down the RNA toward the RNA polymerase. They differ in that rho does not degrade the RNA, whereas Rat1 does so.
WORKED PROBLEMS
The diagram at right represents a sequence of nucleotides surrounding an RNA-coding sequence.
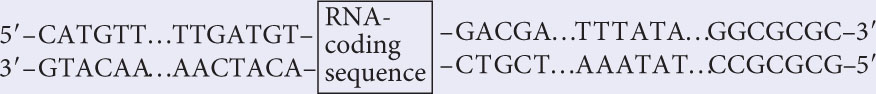
- a. Is the RNA-coding sequence likely to be from a bacterial cell or from a eukaryotic cell? How can you tell?
- b. Which DNA strand will serve as the template strand during the transcription of the RNA-coding sequence?
What information is required in your answer to the problem?
- a. Whether the sequence is likely to be from a bacteria or eukaryotic cell and why.
- b. Which strand is the template strand.
What information is provided to solve the problem?
 The nucleotide sequences of both strands of DNA.
The nucleotide sequences of both strands of DNA. The 5′ and 3′ ends of the strands.
The 5′ and 3′ ends of the strands.
For help with this problem, review:
The Template in Section 13.2, Bacterial Promoters in Section 13.3 and Promoters in Section 13.4.
Solution Steps
- a. Bacterial and eukaryotic cells use the same DNA bases (A, T, G, and C); so the bases themselves provide no clue to the origin of the sequence. The RNA-coding sequence must have a promoter, and bacterial and eukaryotic cells do differ in the consensus sequences found in their promoters; so we should examine the sequences for the presence of familiar consensus sequences. On the bottom strand to the right of the RNA-coding sequence, we find AAATAT, which, written in the conventional manner (5′ on the left), is 5′–TATAAA–3′. This sequence is the TATA box found in most eukaryotic promoters. However, the sequence is also quite similar to the −10 consensus sequence (5′−TATAAT−3′) found in bacterial promoters.Farther to the right on the bottom strand, we also see 5′–GCGCGCC–3′, which is the TFIIB recognition element (BRE, see Figure 13.15) in eukaryotic RNA polymerase II promoters. No similar consensus sequence is found in bacterial promoters; so we can be fairly certain that this sequence is a eukaryotic promoter and an RNA-coding sequence.
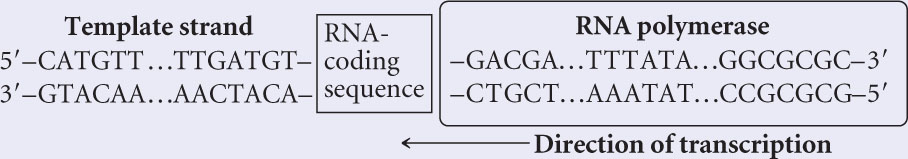
- b. The TATA box and BRE of RNA polymerase II promoters are upstream of the RNA-coding sequences; so RNA polymerase must bind to these sequences and then proceed downstream, transcribing the RNA-coding sequence. Thus RNA polymerase must proceed from right (upstream) to left (downstream). The RNA molecule is always synthesized in the 5′→3′ direction and is antiparallel to the DNA template strand; so the template strand must be read 3′→5′. If the enzyme proceeds from right to left and reads the template in the 3′→5′ direction, the upper strand must be the template, as shown in the diagram below.
Recall: During transcription, the template strand is read 3′→5′.
Suppose that a consensus sequence in the regulatory promoter of a eukaryotic gene that encodes enzyme A were deleted. Which of the following effects would result from this deletion? Explain your reasoning.
- a. Enzyme A would have a different amino acid sequence.
- b. The mRNA for enzyme A would be abnormally short.
- c. Enzyme A would be missing some amino acids.
- d. The mRNA for enzyme A would be transcribed but not translated.
- e. The amount of mRNA transcribed would be affected.
Solution Strategy
What information is required in your answer to the problem?
Selection of the result (a, b, c. d. or e) that would occur when a consensus sequence in the regulatory promoter were deleted.
What information is provided to solve the problem?
The deleted consensus sequence is in the regulatory promoter.
For help with this problem, review:
Section 13.4.
Solution Steps
The correct answer is part e. The regulatory promoter contains binding sites for transcriptional activator proteins. These sequences are not part of the RNA-coding sequence for enzyme A; so the mutation would have no effect on the length or the amino acid sequence of the enzyme, eliminating answers a, b, and c. Transcriptional activator proteins bind to the regulatory promoter and affect the amount of transcription that takes place through interactions with the basal transcription apparatus at the core promoter.