18.4 Transposable Elements Cause Mutations
Transposable elements—sequences that can move about in the genome—are also often a cause of mutations. These mobile DNA elements have been given a variety of names, including transposons, transposable genetic elements, movable genes, controlling elements, and jumping genes. They are found in the genomes of all organisms and are abundant in many: for example, they make up at least 45% of human DNA. Most transposable elements are able to insert at many different locations, relying on mechanisms that are distinct from homologous recombination. Through their movement (transposition), transposable elements often cause mutations, either by inserting into another gene and disrupting it or by promoting DNA rearrangements such as chromosome deletions, duplications, and inversions (see Chapter 8).
General Characteristics of Transposable Elements
There are many different types of transposable elements: some have simple structures, encompassing only those sequences necessary for their own transposition, whereas others have complex structures and encode a number of functions not directly related to transposition. Despite this variation, many transposable elements have certain features in common.
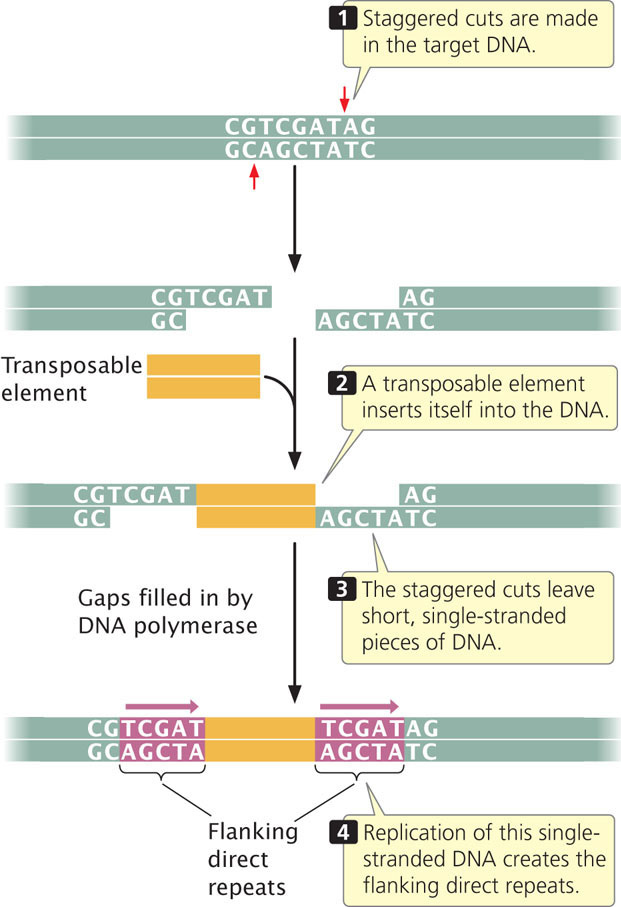
Short flanking direct repeats from 3 to 12 bp long are present on both sides of most transposable elements. The sequences of these repeats vary, but the length is constant for each type of transposable element. These repeats are not a part of a transposable element and do not travel with it. Rather, they are generated in the process of transposition, at the point of insertion. During transposition, flanking repeats are created when staggered cuts are made in the target DNA, as shown in Figure 18.24. The staggered cuts leave short, single-stranded pieces of DNA on either side of the transposable element. Replication of the single-stranded DNA then creates the flanking direct repeats.
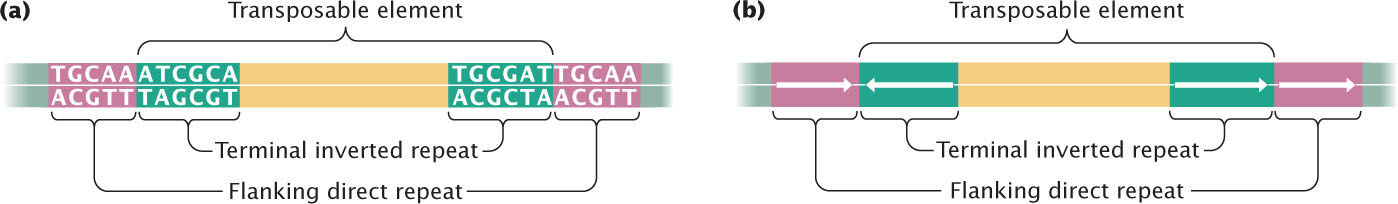
At the ends of many, but not all, transposable elements are terminal inverted repeats—sequences from 9 to 40 bp in length that are inverted complements of one another. For example, the following sequences are inverted repeats:
5′-ACAGTTCAG…CTGAACTGT-3′
3′-TGTCAAGTC…GACTTGACA-5′
On the same strand, the two sequences are not simple inversions, as their name might imply; rather, they are both inverted and complementary. (Notice that the sequence from left to right in the top strand is the same as the sequence from right to left in the bottom strand.) Terminal inverted repeats are recognized by enzymes that catalyze transposition and are required for transposition to take place. Figure 18.25 summarizes the general characteristics of transposable elements.  TRY PROBLEM 33
TRY PROBLEM 33
CONCEPTS
Transposable elements are mobile DNA sequences that often cause mutations. There are many different types of transposable elements; most generate short flanking direct repeats at the target sites as they insert. Many transposable elements also possess short terminal inverted repeats.
 CONCEPT CHECK 5
CONCEPT CHECK 5How are flanking direct repeats created in transposition?
Transposition
As mentioned above, transposition is the movement of a transposable element from one location to another. Several different mechanisms are used for transposition in both prokaryotic and eukaryotic cells. Nevertheless, all types of transposition have several features in common: (1) staggered breaks are made in the target DNA (see Figure 18.24); (2) the transposable element is joined to single-stranded ends of the target DNA; and (3) DNA is replicated at the single-strand gaps. A transposase enzyme, often encoded by the transposable element, is used to make the staggered breaks in DNA and to integrate the transposable element into a new site, both of which enable the transposable element to move.
Some transposable elements transpose as DNA and are referred to as DNA transposons (also called Class II transposable elements). Other transposable elements transpose through an RNA intermediate. In this case, RNA is transcribed from the transposable element (DNA) and is then copied back into DNA by a special enzyme called reverse transcriptase. Elements that transpose through an RNA intermediate are called retrotransposons (also called Class I transposons). Most transposable elements found in bacteria are DNA transposons. Both DNA transposons and retrotransposons are found in eukaryotes, although retrotransposons are more common.
Among DNA transposons, transposition may be replicative or nonreplicative. In replicative transposition (also called copy-and-paste transposition), a new copy of the transposable element is introduced at a new site while the old copy remains behind at the original site, and so the number of copies of the transposable element increases as a result of transposition. In nonreplicative transposition (cut-and-paste transposition), the transposable element excises from the old site and inserts at a new site without any increase in the number of its copies. Nonreplicative transposition requires the replication of only the few nucleotides that constitute the direct repeats. Retrotransposons use replicative transposition only.
Control of Transposition
Many organisms limit transposition by methylating the DNA in regions where transposons are common. DNA methylation usually suppresses transcription (see Chapter 17), preventing the production of the transposase enzyme necessary for transposition. Alterations of chromatin structure also are used to prevent the transcription of transposons. In other cases, translation of the transposase mRNA is controlled. Some animals use small RNA molecules called Piwi-interacting RNAs (piRNAs, see Chapter 14) to silence transposons; piRNAs combine with Piwi proteins and inhibit the expression of transposons sequences.
Transposition in Humans
About 45% of the human genome is comprised of sequences that are related to transposable elements, mostly retrotransposons (some research suggests that almost two-thirds of the human genome consists of transposable elements). Researchers previously assumed that most of these transposable elements are inactive and that little transposition occurs today, although it clearly took place extensively during past evolution. More recently, however, researchers have begun to map copies of transposons across the genome and have discovered that people often differ in the number and location of transposons. This suggests that recent transpositions are more common than previously thought. The L1 transposon, for example, is estimated to undergo one transposition event about every 100 human births.
Research has also demonstrated that some cancer cells have elevated levels of transposition, probably because patterns of DNA methylation that normally inhibit transposition are disrupted in these cells.
CONCEPTS
Transposition may take place through DNA or an RNA intermediate. In replicative transposition, a new copy of the transposable element inserts in a new location and the old copy stays behind; in nonreplicative transposition, the old copy excises from the old site and moves to a new site. Transposition through an RNA intermediate requires reverse transcription to integrate into the target site. Many cells regulate transposition by a variety of mechanisms.
The Mutagenic Effects of Transposition
Because transposable elements can insert into other genes and disrupt their function, transposition is generally mutagenic. In fact, more than half of all spontaneously occurring mutations in Drosophila result from the insertion of a transposable element in or near a functional gene.
A number of cases of human genetic disease have been traced to the insertion of a transposable genetic element into a vital gene. For example, insertion of the L1 transposable element into the gene for blood clotting factor VIII has caused hemophilia. Although most mutations resulting from transposition are detrimental, transposition may occasionally activate a gene or change the phenotype of the cell in a beneficial way. For instance, bacterial transposable elements sometimes carry genes that encode antibiotic resistance, and several transposable elements have created mutations that confer insecticide resistance in insects.
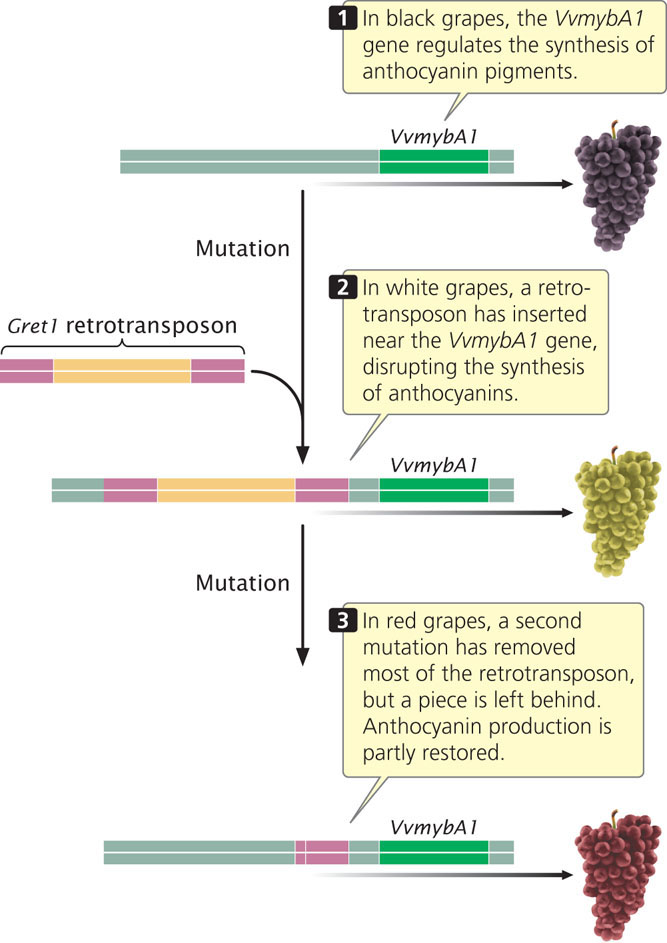
A dramatic example of the mutagenic effect of transposable elements is seen in the color of grapes, which come in black, red, and white varieties (Figure 18.26). Black and red grapes result from the production of red pigments (anthocyanins) in the skin, which are lacking in white grapes. White grapes resulted from a mutation in black grapes that turned off the production of anthocyanin pigments. This mutation consisted of the insertion of a 10,422-bp retrotransposon called Gret1 near a gene that promotes the production of anthocyanins. The Gret1 retrotransposon apparently disrupted sequences that regulate the gene, effectively shutting down pigment production and producing a white grape with no anthocyanins. Interestingly, red grapes resulted from a second mutation taking place in the white grapes (see Figure 18.26). This mutation (probably resulting from faulty recombination) removed most but not all of the retrotransposons, switching pigment production back on, but not as intensely as in the original black grapes.
Because transposition entails the exchange of DNA sequences and recombination, it often leads to DNA rearrangements. Homologous recombination between multiple copies of transposons can lead to duplications, deletions, and inversions, as shown in Figure 18.27. The Bar mutation in Drosophila (see Figure 8.6) is a tandem duplication thought to have arisen through homologous recombination between two copies of a transposable element present in different locations on the X chromosome. Similarly, recombination between copies of the transposable element Rider caused a duplication that results in elongated fruit in tomatoes.
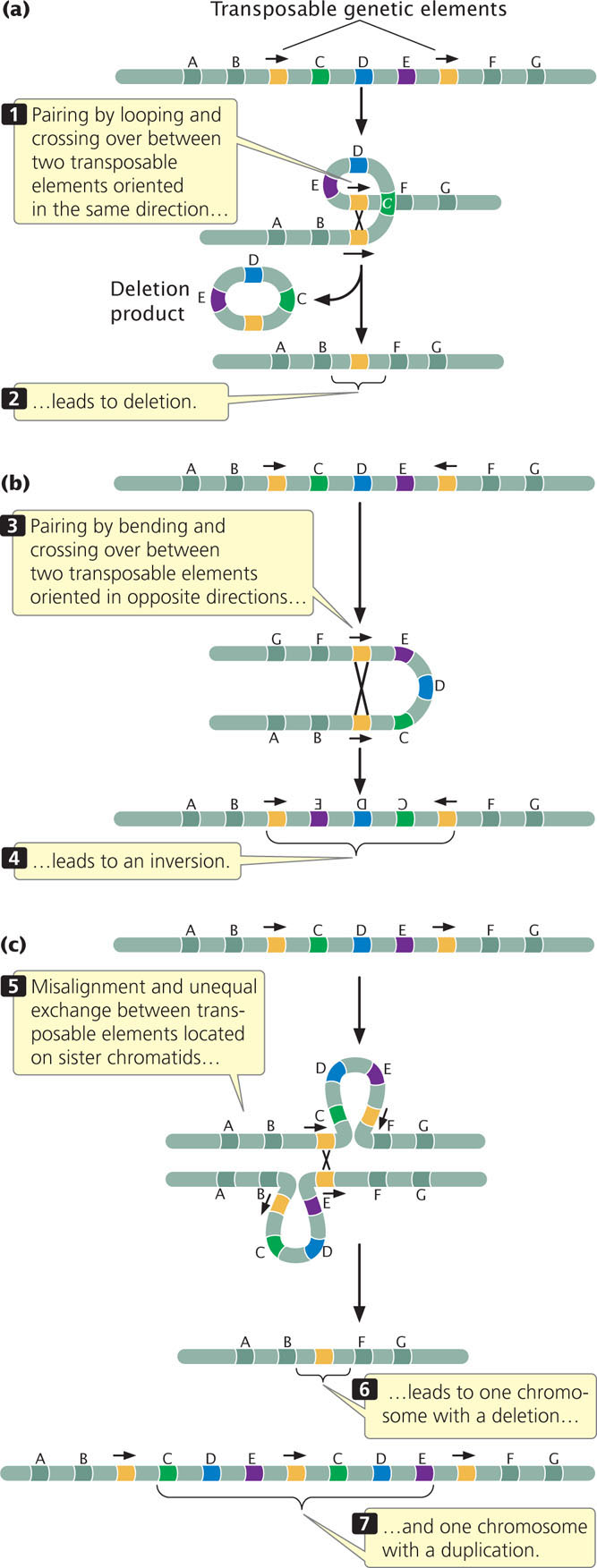
DNA rearrangements can also be caused by the excision of transposable elements in a cut-and-paste transposition. If the broken DNA is not repaired properly, a chromosome rearrangement can be generated.
Because most transposable elements insert randomly into DNA sequences, they provide researchers with a powerful tool for inducing mutations throughout the genome, allowing them to determine the functions of genes, study genetic phenomena, and map genes. Furthermore, because the transposable element being used has a known sequence, it can serve as a “tag” for locating the gene in which the mutation has occurred. For example, researchers engineered a transposable element named Sleeping Beauty to induce mutations in mice and used it to search for genes that cause cancer. Sleeping Beauty was introduced into a strain of mice that produce the transposase needed for transposition, and the transposable element inserted randomly into different locations in the genome. Occasionally, it inserted into a gene that protects against cancer and destroyed its function. By looking for the location of the Sleeping Beauty sequence in the DNA from the tumor cells that subsequently developed, geneticists identified a number of genes that protect against cancer.
Bacteria and eukaryotic organisms possess a number of different types of transposable elements, the structures of which vary extensively. In the next two sections, we consider the structure and types of transposable elements in bacteria and eukaryotes.
CONCEPTS
Transposable elements frequently cause mutations and DNA rearrangements.
 CONCEPT CHECK 6
CONCEPT CHECK 6Briefly explain how transposition causes mutations and chromosome rearrangements.
Transposable Elements in Bacteria
The DNA transposons found in bacteria (there are no retrotransposons in bacteria) constitute two major groups: (1) simple transposable elements, called insertion sequences, that carry only the information required for movement and (2) more-complex transposable elements, called composite transposons, which contain DNA sequences not directly related to transposition.
Insertion Sequences
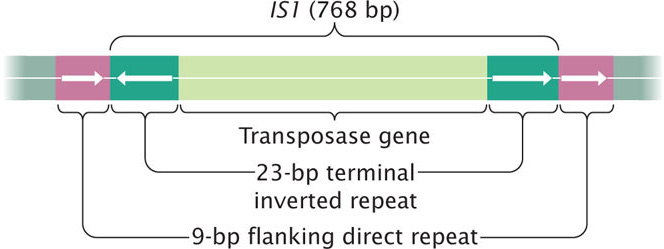
The simplest type of transposable element in bacterial chromosomes and plasmids is an insertion sequence (IS). This type of element carries only the genetic information necessary for its movement. Insertion sequences are common constituents of bacteria; they can also infect plasmids and viruses and, in this way, can be passed from one cell to another. Geneticists designate each type of insertion sequence with IS followed by an identifying number. For example, IS1 is a common insertion sequence found in E. coli.
A number of different insertion sequences have been found in bacteria. They are typically from 800 to 2000 bp in length and possess the two hallmarks of transposable elements: terminal inverted repeats and the generation of flanking direct repeats at the site of insertion. Most insertion sequences contain one or two genes that encode transposase. IS1, a typical insertion sequence, is shown in Figure 18.28.  TRY PROBLEM 18
TRY PROBLEM 18
Composite Transposons
Any segment of DNA that becomes flanked by two copies of an insertion sequence may itself transpose and is called a composite transposon. Each composite transposon is designated by the abbreviation Tn, followed by a number. The composite transposon Tn10, for example, consists of about 9300 bp that carries a gene for tetracycline resistance between two IS10 insertion sequences (Figure 18.29). The insertion sequences have terminal inverted repeats, so the composite transposon also ends in inverted repeats. Composite transposons also generate flanking direct repeats at their sites of insertion (see Figure 18.29). The insertion sequences at the ends of a composite transposon may be in the same orientation or they may be inverted relative to one other (as in Tn10).
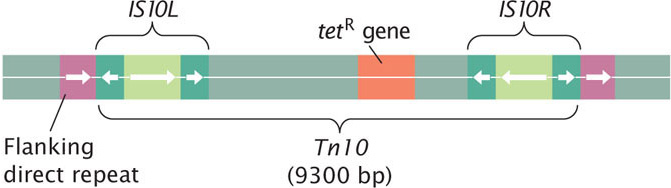
The insertion sequences at the ends of a composite transposon are responsible for transposition. The DNA between the insertion sequences is not required for movement and may carry additional information (such as antibiotic resistance). Presumably, composite transposons evolve when one insertion sequence transposes to a location close to another of the same type. The transposase produced by one of the insertion sequences catalyzes the transposition of both insertions sequences, allowing them to move together and carry along the DNA that lies between them. In some composite transposons (such as Tn10) one of the insertion sequences may be defective, so its movement depends on the transposase produced by the other.
Noncomposite Transposons
Some transposable elements in bacteria lack insertion sequences and are referred to as noncomposite transposons. Noncomposite transposons possess a gene for transposase and have inverted repeats at their ends. For instance, the noncomposite transposon Tn3 carries genes for transposase and resolvase (an enzyme that functions in recombination), plus a gene that encodes the enzyme β-lactamase, which provides resistance to the antibiotic ampicillin.
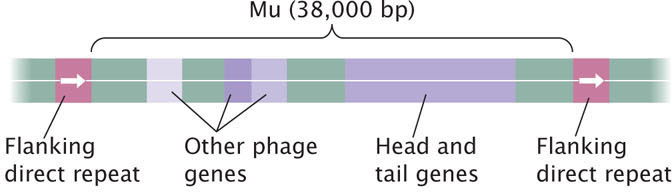
A few bacteriophage genomes reproduce by transposition and use transposition to insert themselves into a bacterial chromosome in their lysogenic cycle; the best studied transposing bacteriophage is Mu (Figure 18.30). Although Mu does not possess terminal inverted repeats, it does generate short (5-bp) flanking direct repeats when it inserts randomly into DNA. Mu replicates through transposition and causes mutations at the site of insertion, properties characteristic of transposable elements.
CONCEPTS
Insertion sequences are prokaryotic transposable elements that carry only the information needed for transposition. A composite transposon consists of two insertion sequences plus intervening DNA. Noncomposite transposons in bacteria lack insertion sequences but have terminal inverted repeats and carry information not related to transposition. All of these transposable elements generate flanking direct repeats at their points of insertion.
 CONCEPT CHECK 7
CONCEPT CHECK 7Which type of transposable element possesses terminal inverted repeats?
- Insertion sequence.
- Composite transposons.
- Noncomposite transposon Tn3.
- All the above.
Transposable Elements in Eukaryotes
Eukaryotic transposable elements can be divided into two groups. One group is structurally similar to transposable elements in bacteria, typically ending in short inverted repeats and transposing as DNA: examples include the P elements in Drosophila and the Ac and Ds elements in maize (corn). The other group comprises retrotransposons; they use RNA intermediates, and many are similar in structure and movement to retroviruses (see Chapter 9). On the basis of their structure, function, and genomic sequences, some retrotransposons are clearly evolutionarily related to retroviruses. Although their mechanism of movement is fundamentally different from that of other transposable elements, retrotransposons also generate direct repeats at the point of insertion. Retrotransposons include the Ty elements in yeast, the copia elements in Drosophila, and the Alu sequences in humans.
Ac and Ds Elements in Maize
Transposable elements were first identified in maize more than 50 years ago by Barbara McClintock (Figure 18.31). McClintock spent much of her long career studying their properties, and her work stands among the landmark discoveries of genetics. Her results, however, were misunderstood and ignored for many years. Not until molecular techniques were developed in the late 1960s and 1970s did the importance of transposable elements become widely accepted. The significance of McClintock’s early discoveries was finally recognized in 1983, when she was awarded the Nobel Prize in physiology or medicine.

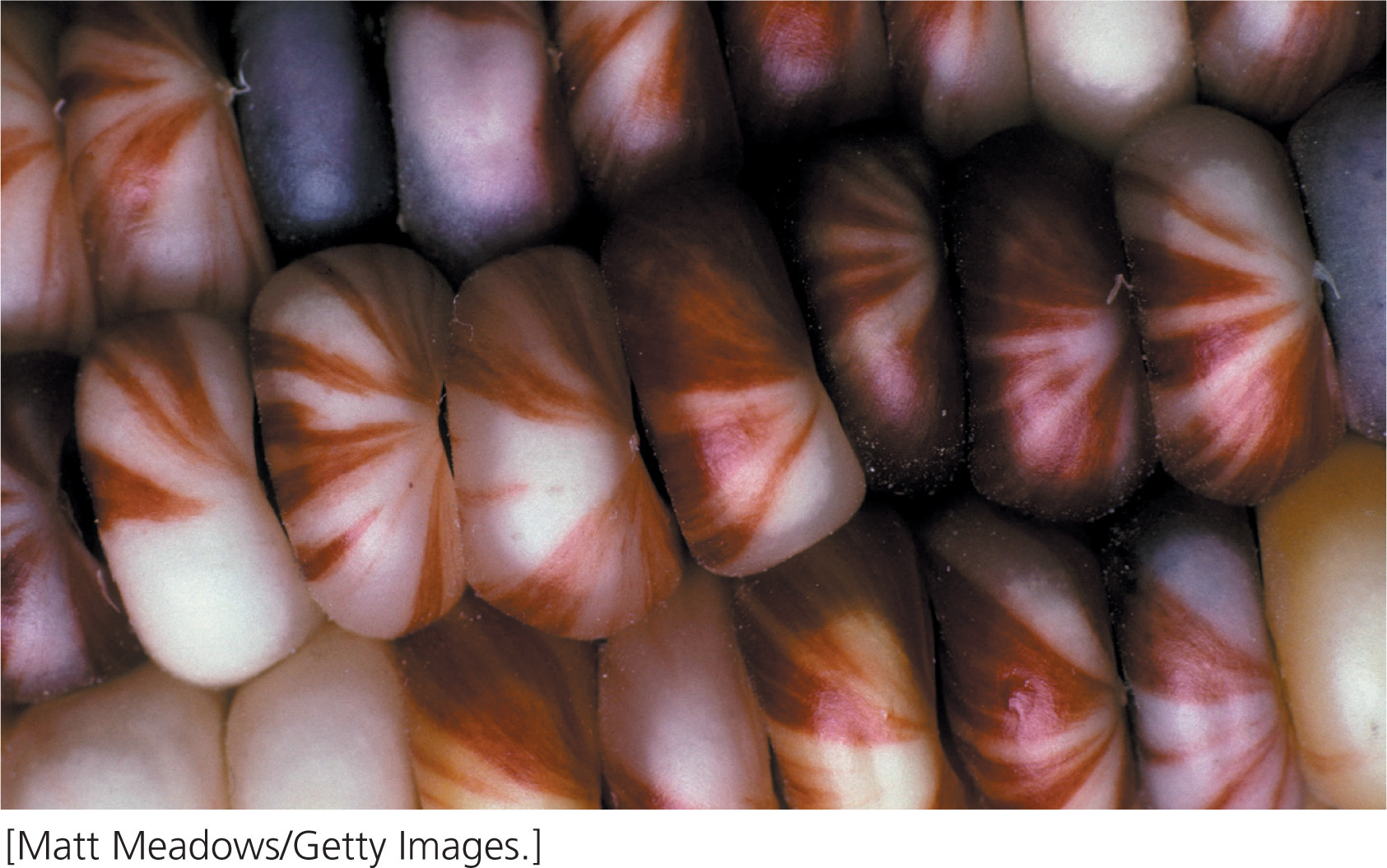
McClintock’s discovery of transposable elements had its genesis in the early work of Rollins A. Emerson on the maize genes that caused variegated (multicolored) kernels. Most corn kernels are either wholly pigmented or colorless (yellow), but Emerson noted that some yellow kernels had spots or streaks of color (Figure 18.32). He proposed that these kernels resulted from an unstable mutation: a mutation in the wild-type gene for pigment produced a colorless kernel but, in some cells, the mutation reverted back to the wild type, causing a spot of pigment. However, Emerson didn’t know why these mutations were unstable.
McClintock discovered that the cause of the unstable mutation was a gene that moved. She noticed that chromosome breakage in maize often occurred at a gene that she called Dissociation (Ds) but only if another gene, the Activator (Ac), also was present. Occasionally, the genes moved together to a different chromosomal location. McClintock called these moving genes controlling elements, because they controlled the expression of other genes.
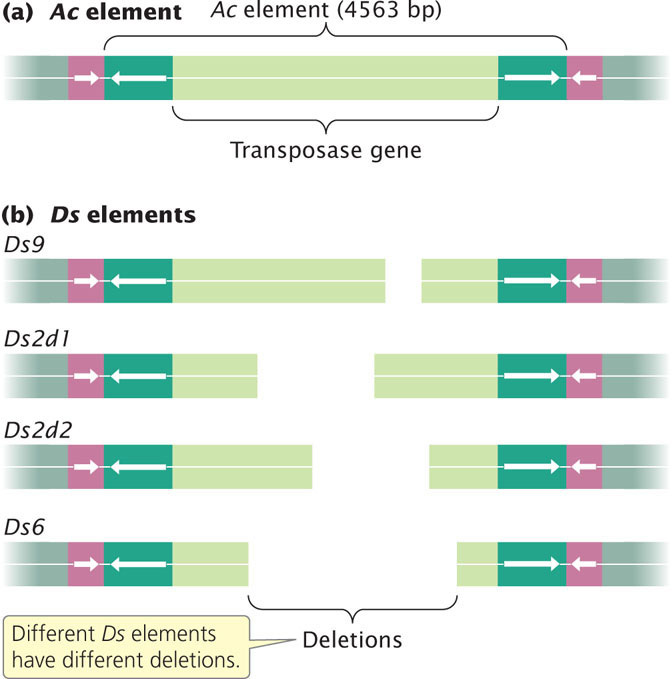
Since the significance of McClintock’s work was recognized, Ac and Ds elements in maize have been examined in detail. They are DNA transposons that possess terminal inverted repeats and generate flanking direct repeats at the points of insertion (Figure 18.33a). Each Ac element contains a single gene that encodes a transposase enzyme. Thus Ac elements are autonomous—able to transpose. Ds elements are Ac elements with one or more deletions that have inactivated the transposase gene (Figure 18.33b). Unable to transpose on their own (nonautonomous), Ds elements can transpose in the presence of Ac elements because they still possess terminal inverted repeats recognized by Ac transposase.
Each kernel in an ear of corn is an individual offspring, originating as an ovule fertilized by a pollen grain. A kernel’s pigment pattern is determined by several loci. A pigment-encoding allele at one of these loci can be designated C, and an allele at the same locus that does not confer pigment can be designated c. A kernel with genotype cc will be colorless—that is, yellow or white (Figure 18.34a); a kernel with genotype CC or Cc will produce pigment and be purple (Figure 18.34b).
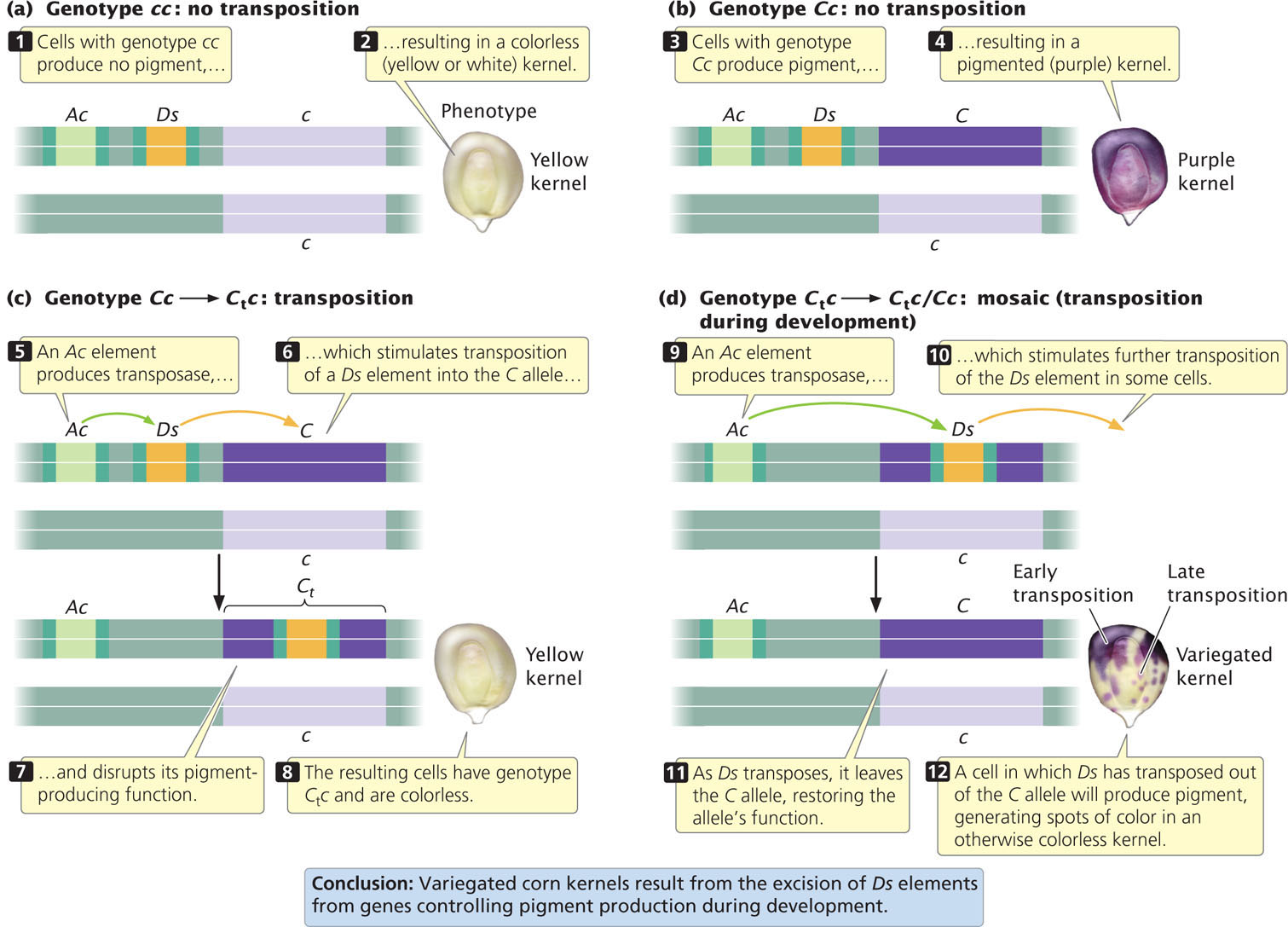
A Ds element, transposing under the influence of a nearby Ac element, may insert into the C allele, destroying its ability to produce pigment (Figure 18.34c). An allele inactivated by a transposable element is designated by a subscript “t”; so, in this case, the allele would be Ct.
If a kernel is initially heterozygous with genotype Cc, then after the transposition of Ds into the C allele, the kernel cell has genotype Ctc. This kernel will be colorless (white or yellow), because neither the Ct allele nor the c allele confers pigment. As the original one-celled maize embryo develops and divides by mitosis, additional transpositions may take place in some cells. In any cell in which the transposable element excises from the Ct allele and moves to a new location, the C allele may be rendered functional again: all cells derived from those in which this event has taken place will have the genotype Cc and be purple. The presence of these pigmented cells, surrounded by the colorless (Ctc) cells, produces a purple spot or streak (called a sector) in the otherwise yellow kernel (Figure 18.34d). The size of the sector varies, depending on when the excision of the transposable element from the Ct allele takes place. If excision is early in development, then many cells will contain the functional C allele and the pigmented sector will be large; if excision is late in development, few cells will have the functional C allele and the pigmented sector will be small.  TRY PROBLEM 42
TRY PROBLEM 42
Transposable Elements in Drosophila
A number of different transposable elements are found in Drosophila. One family of Drosophila transposable elements comprises the P elements. Most functional P elements are about 2900 bp long, although shorter P elements containing deletions also exist. Each P element possesses terminal inverted repeats and generates flanking direct repeats at the site of insertion. Like transposable elements in bacteria, P elements are DNA transposons. Each element encodes both a transposase and a repressor of transposition.
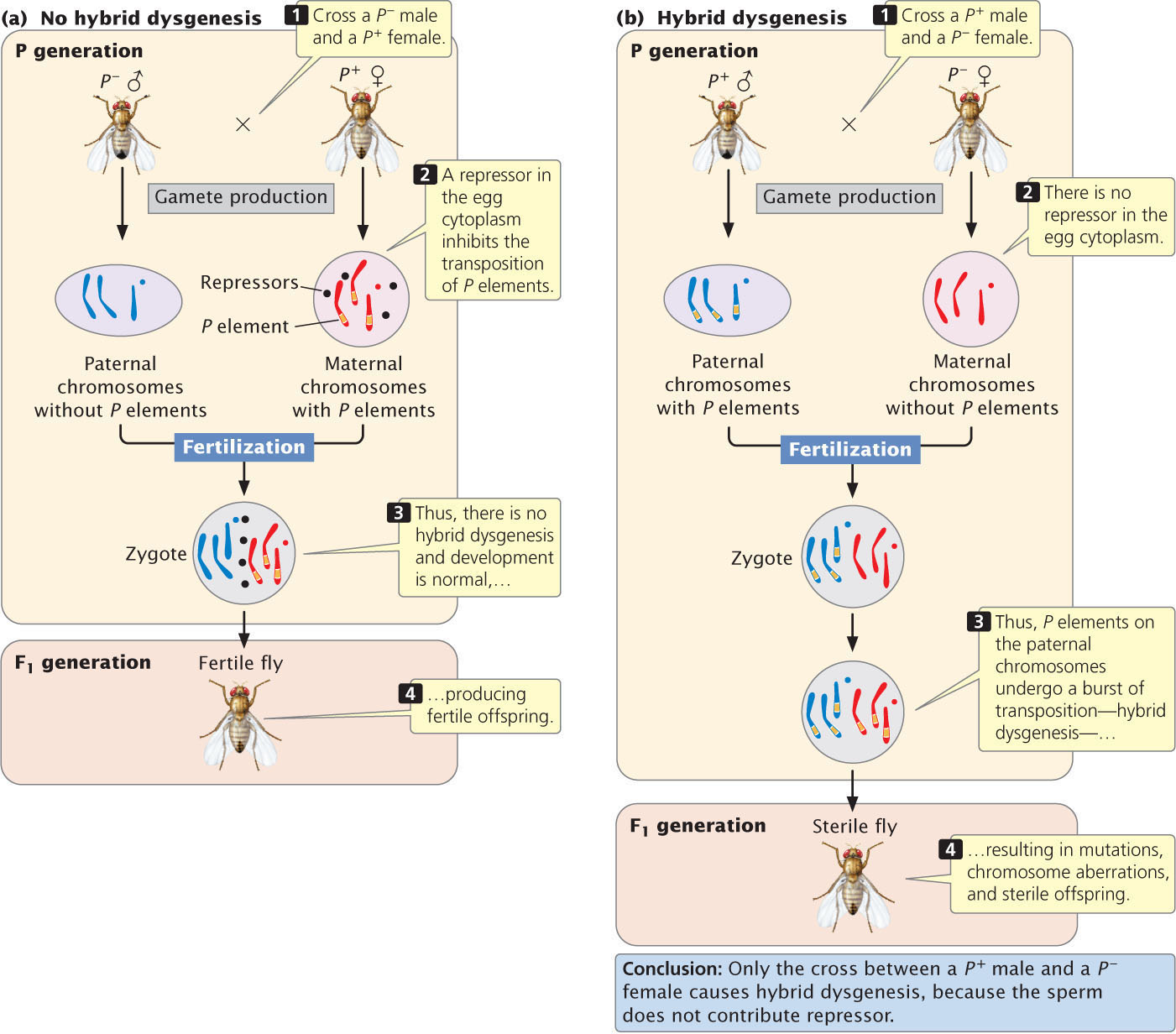
The role of this repressor in controlling transposition is demonstrated dramatically in hybrid dysgenesis, which is the sudden appearance of numerous mutations, chromosome aberrations, and sterility in the offspring of a cross between a P+ male fly (with P elements) and a P− female fly(without them). The reciprocal cross between a P+ female and a P− male produces normal offspring.
Hybrid dysgenesis arises from a burst of transposition when P elements are introduced into a cell that does not possess them. In a cell that contains P elements, a repressor in the cytoplasm inhibits transposition. When a P+ female produces eggs, the repressor protein is incorporated into the egg cytoplasm, which prevents further transposition in the embryo and thus prevents mutations from arising. The resulting offspring are fertile as adults (Figure 18.35a). However, a P− female does not produce the repressor protein; so none is stored in the cytoplasm of her eggs. Sperm contain little or no cytoplasm, so a P+ male does not contribute the repressor protein to his offspring. When eggs from a P− female are fertilized by sperm from a P+ male, the absence of repression allows the P elements contributed by the sperm to undergo rapid transposition in the embryo, causing hybrid dysgenesis (Figure 18.35b).
Hybrid dysgenesis and P elements have attracted geneticists’ attention because P elements appear to have arisen within populations of Drosophila melanogaster within the past 50 years and may play a role in the species evolution. Other species of Drosophila lack P elements, which are also completely absent from laboratory strains of D. melanogaster originally collected from the wild prior to the 1960s. However, today most wild populations of the species have P elements. Laboratory strains collected during the 1970s are mixed: some have P elements and some do not. This suggests that P elements arose sometime during the past century and spread quickly throughout all wild populations of D. melanogaster. Because crosses between males with P elements and females without them cause sterility, P elements and similar transposable elements have the potential to serve as reproductive isolating mechanisms between populations and may play a role in bringing about speciation (see Chapter 26). These observations support the idea that transposable elements play important roles in evolution.  TRY PROBLEM 38
TRY PROBLEM 38
Transposable Elements in Humans
One of the most common transposable elements in the human genome is Alu. Every human cell contains more than 1 million related but not identical copies of Alu in its chromosomes. Alu sequences are similar to the gene that encodes the 7S RNA molecule, which transports newly synthesized proteins across the endoplasmic reticulum. Alu sequences create short flanking direct repeats when they insert into DNA and have characteristics that suggest that they have transposed through an RNA intermediate.
Alu belongs to a class of repetitive sequences found frequently in mammalian and some other genomes. These sequences are collectively referred to as short interspersed elements (SINEs) and constitute about 11% of the human genome. Most SINEs are copies of transposable elements that have been shortened at the 5′ end, probably because the reverse-transcription process used in their transposition terminated before the entire sequence was copied. SINEs have been identified as the cause of mutations in more than 20 cases of human genetic disease.
The human genome also has many transposons classified as long interspersed elements (LINEs), which are somewhat more similar in structure to retroviruses. Like SINEs, most LINEs in the human genome have been shortened at the 5′ end. The longest LINEs are usually about 6000 bp but, because most copies are shortened, the average LINE is only about 900 bp. There are approximately 900,000 copies of LINEs in the human genome, collectively constituting 21% of the total human DNA.
CONCEPTS
A great variety of transposable elements exist in eukaryotes. Some resemble transposable elements in prokaryotes, having terminal inverted repeats, and transpose as DNA. Others are retrotransposons with long direct repeats at their ends and transpose through an RNA intermediate.
 CONCEPT CHECK 8
CONCEPT CHECK 8Hybrid dysgenesis results when
- a male fly with P elements (P+) mates with a female fly that lacks P elements (P−).
- a P− male mates with a P+ female.
- a P+ male mates with a P+ female.
- a P− male mates with a P− female.
CONNECTING CONCEPTS
Now that we have looked at some examples of transposable elements, let’s review their major types (Table 18.4).
| Structure | Genes Encoded | Transposition | Examples | |
|---|---|---|---|---|
| Class I (retrotransposon) | Long terminal direct repeats; short flanking direct repeats at target site | Reverse-transcriptase gene (and sometimes others) | By RNA intermediate | Ty (yeast) copia (Drosophila) Alu (human) |
| Class II | Short terminal inverted repeats; short flanking direct repeats at target site | Transposase gene (and sometimes others) | Through DNA (replicative or nonreplicative) | IS1 (E. coli) Tn3 (E. coli) Ac, Ds (maize) P elements (Drosophila) |
Transposable elements can be divided into two major classes on the basis of structure and movement. Class I comprises the retrotransposons, which possess terminal direct repeats and transpose through RNA intermediates. They generate flanking direct repeats at their points of insertion when they transpose into DNA. Retrotransposons do not encode transposase, but some types are similar in structure to retroviruses and carry sequences that produce reverse transcriptase. Transposition takes place when transcription produces an RNA intermediate, which is then transcribed into DNA by reverse transcriptase and inserted into the target site. Examples of retrotransposons include Ty elements in yeast and Alu sequences in humans. Retrotransposons are not found in prokaryotes.
Class II consists of DNA transposons that possess terminal inverted repeats and transpose as DNA. Like Class I transposons, they all generate flanking direct repeats at their points of insertion into DNA. Unlike Class I transposons, all active forms of Class II transposable elements encode transposase, which is required for their movement. Some also encode resolvase, repressors, and other proteins. Their transposition may be replicative or nonreplicative, but they never use RNA intermediates. Examples of transposable elements in this class include insertion sequences and all complex transposons in bacteria, Ac and Ds elements in maize, and P elements in Drosophila.
Transposable Elements Have Played an Important Role in Genome Evolution
Transposable elements have clearly played an important role in shaping the genomes of many organisms. Much of the tremendous variation in genome size found among eukaryotic organisms is due to differences in numbers of transposable elements. Approximately 45% of the human genome consists of remnants of transposable elements and about 50% of all spontaneous mutations in Drosophila are due to transposition. Homologous recombination between copies of transposable elements has been an important force in producing gene duplications and other chromosome rearrangements. Furthermore, some transposable elements may carry extra DNA with them when they transpose to a new site, providing the potential to move DNA sequences that regulate genes to new sites, where they may alter the expression of genes.
Transposable Elements As Genomic Parasites
As we have seen, many transposable elements leave a copy behind when they transpose to a new location (copy-and-paste transposition) and therefore increase in number within a genome with the passage of time. This ability to replicate and spread means that many transposable elements may serve no purpose for the cell; they exist simply because they are capable of replicating and spreading. The insertion of transposable elements into a gene will often destroy its function, with harmful consequences for the cell. Furthermore, the time and energy required to replicate large numbers of transposable elements are likely to place a metabolic burden on the cell. Thus, transposable elements can be thought of as genomic parasites that provide no benefit to the cell and may even be harmful.
Domestication Of Transposable Elements
Although many transposable elements may be genomic parasites, some have clearly evolved to serve useful purposes for their host cells. These transposons are sometimes referred to as domesticated, implying that their parasitic tendencies have been replaced by properties useful to the cell. For example, the mechanism that generates antibody diversity in the immune systems of vertebrates (see Chapter 22) probably evolved from a transposable element. Immune cells called lymphocytes have the ability to unite several DNA segments that encode antigen-recognition proteins. This mechanism may have arisen from a transposable element that inserted into the germ line of a vertebrate ancestor some 450 million years ago.
Transposable elements have also been played an important role in the evolution of maize. Maize (modern corn) was domesticated from teosinte in Central America more than 8000 years ago. One of the important genetic differences between teosinte and corn involves a gene called tb1, which encodes a transcriptional regulator that represses the growth of side branches. In corn, transcription of tb1 is elevated compared with that of teosinte, with the result that modern corn is more upright and less branched than teosinte. Recent research has demonstrated that the elevated transcription of tb1 in corn is the result of a transposable element called Hopscotch that inserted into regulatory sequences that control tb1 transcription. This insertion was probably present in teosinte as a variant and was selected by humans during the domestication of corn because it produced a more desirable plant shape. Transposable elements may also play a role in speciation, the process by which new species arise (see earlier section on hybrid dysgenesis).
CONCEPTS
Many transposable elements appear to be genomic parasites, existing in large numbers because of their ability to efficiently increase in copy number. Increases in copy number of transposable elements have contributed to the large size of many eukaryotic genomes. In several cases, transposable elements have been adopted for specific cellular functions.