APPLICATION QUESTIONS AND PROBLEMS
Question 15
*15.An RNA molecule has the following percentages of bases: A = 23%, U = 42%, C = 21%, and G = 14%.
Is this RNA single stranded or double stranded? How can you tell?
What would be the percentages of bases in the template strand of the DNA that contains the gene for this RNA?
Question 16
*16.The following diagram represents DNA that is part of the RNA-
|
5′—ATAGGCGATGCCA— |
|
|
3′—TATCCGCTACGGT— |
← Template strand |
Question 17
17.For the RNA molecule shown in Figure 10.1a, write out the sequence of bases on the template and nontemplate strands of DNA from which this RNA is transcribed. Label the 5′ and 3′ ends of each strand.
Question 18
18.The following sequence of nucleotides is found in a single-
ATTGCCAGATCATCCCAATAGAT
Assume that RNA polymerase proceeds along this template from left to right.
Which end of the DNA template is 5′ and which end is 3′?
Give the sequence and identify the 5′ and 3′ ends of the RNA copied from this template.
Question 19
19.List at least five properties that DNA polymerases and RNA polymerases have in common. List at least three differences.
Question 20
20.Most RNA molecules have three phosphate groups at the 5′ end, but DNA molecules never do. Explain this difference.
Question 21
21.A strain of bacteria possesses a temperature-
Question 22
22.On Figure 10.5, indicate the locations of the promoters and terminators for genes a, b, and c.
Question 23
23.The following diagram represents a transcription unit on a DNA molecule.
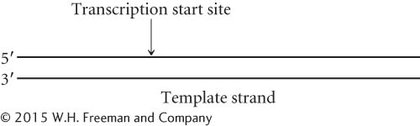
Assume that this DNA molecule is from a bacterial cell. Label the approximate locations of the promoter and terminator for this transcription unit.
Question 24
24.The following diagram represents one of the Christmas-
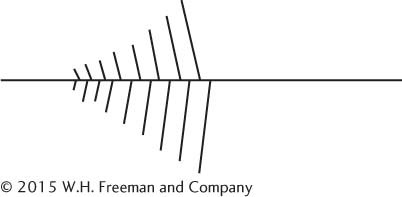
DNA molecule
5′ and 3′ ends of the template strand of DNA
At least one RNA molecule
5′ and 3′ ends of at least one RNA molecule
Direction of movement of the transcription apparatus on the DNA molecule
Approximate location of the promoter
Possible location of a terminator
Upstream and downstream directions
Molecules of RNA polymerase (use dots to represent these molecules)
Question 25
25.Provide the consensus sequence for the first three actual sequences shown in Figure 10.9.
Question 26
*26.Write the consensus sequence for the following set of nucleotide sequences.
AGGAGTT
AGCTATT
TGCAATA
ACGAAAA
TCCTAAT
TGCAATT
Question 27
*27.What would be the most likely effect of a mutation at the following locations in an E. coli gene?
−8
−35
−20
Start site of transcription
Question 28
*28.Duchenne muscular dystrophy is caused by a mutation in a gene that encompasses more than 2 million nucleotides and specifies a protein called dystrophin. However, less than 1% of the gene actually encodes the amino acids in the dystrophin protein. On the basis of what you now know about gene structure and RNA processing in eukaryotic cells, provide a possible explanation for the large size of the dystrophin gene.
Question 29
29.What would be the most likely effect of moving the AAUAAA consensus sequence shown in Figure 10.19 ten nucleotides upstream?
Question 30
30.Suppose that a mutation occurs in the middle of a large intron of a gene encoding a protein. What will the most likely effect of the mutation be on the amino acid sequence of that protein? Explain your answer.
Question 31
31.A geneticist isolates a gene that contains eight exons. He then isolates the mature mRNA produced by this gene. After making the DNA single stranded, he mixes the single-
Question 32
*32.Draw a typical eukaryotic gene and the pre-
5′ untranslated region
Promoter
AAUAAA consensus sequence
Transcription start site
3′ untranslated region
Introns
Exons
Poly(A) tail
5′ cap
Question 33
33.How would the deletion of the Shine–Dalgarno sequence affect a bacterial mRNA?
Question 34
34.A geneticist discovers that two different proteins are encoded by the same gene. One protein has 56 amino acids, and the other has 82 amino acids. Provide a possible explanation for how the same gene can encode both of these proteins.
Question 35
35. In the early 1990s, Carolyn Napoli and her colleagues were attempting to genetically engineer a variety of petunias with dark purple petals by introducing numerous copies of a gene that encodes purple petals (C. Napoli, C. Lemieux, and R. Jorgensen. 1990. Plant Cell 2:279–289). Their thinking was that extra copies of the gene would cause more purple pigment to be produced and would result in a petunia with an even darker hue of purple. However, much to their surprise, many of the plants carrying extra copies of the purple gene were completely white or had only patches of color. Molecular analysis revealed that the level of the mRNA produced by the purple gene was reduced 50-
In the early 1990s, Carolyn Napoli and her colleagues were attempting to genetically engineer a variety of petunias with dark purple petals by introducing numerous copies of a gene that encodes purple petals (C. Napoli, C. Lemieux, and R. Jorgensen. 1990. Plant Cell 2:279–289). Their thinking was that extra copies of the gene would cause more purple pigment to be produced and would result in a petunia with an even darker hue of purple. However, much to their surprise, many of the plants carrying extra copies of the purple gene were completely white or had only patches of color. Molecular analysis revealed that the level of the mRNA produced by the purple gene was reduced 50-
