Gene Regulation by RNA Processing and Degradation
In bacteria, transcription and translation take place simultaneously. In eukaryotes, transcription takes place in the nucleus and the pre-
GENE REGULATION THROUGH RNA PROCESSING Alternative splicing allows a pre-
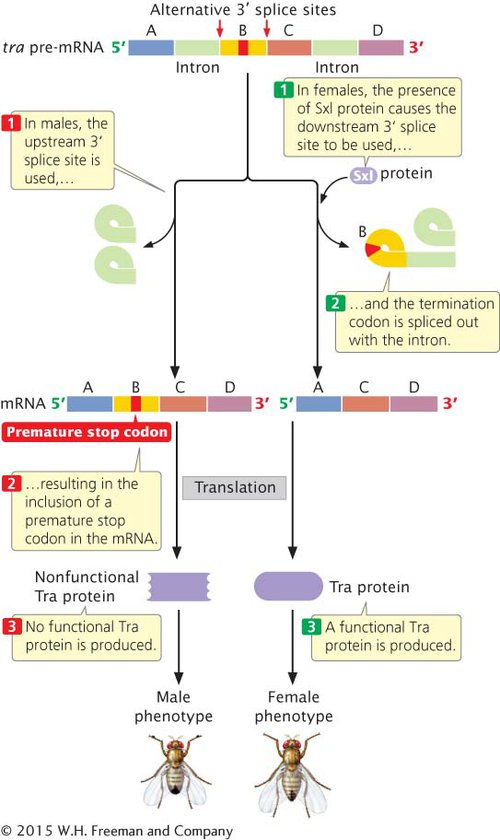
An example of regulation by alternative mRNA splicing is sex determination in fruit flies (see Sex Determination in Drosophila melanogaster in Section 4.1). Sex differentiation in Drosophila arises from a cascade of gene regulation. In XX fly embryos, a female-
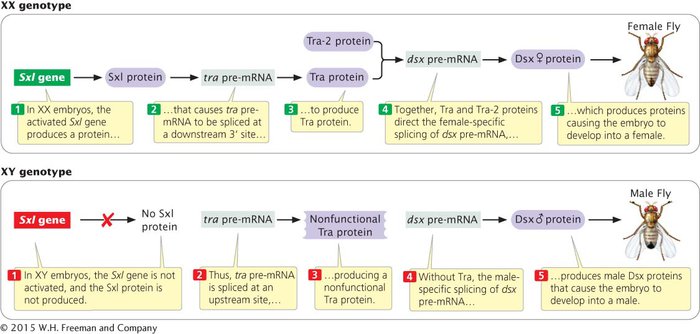
In XY fly embryos, the promoter that transcribes the Sxl gene in females is inactive, so no Sxl protein is produced. In the absence of Sxl protein, tra pre-
CONCEPTS
Eukaryotic genes can be regulated through the control of mRNA processing. The selection of alternative splice sites leads to the production of different proteins.
THE DEGRADATION OF RNA The amount of a protein that is synthesized depends on the amount of corresponding mRNA available for translation. The amount of available mRNA, in turn, depends on both the rate of mRNA synthesis and the rate of mRNA degradation. Eukaryotic mRNAs are generally more stable than bacterial mRNAs, which typically last only a few minutes before being degraded. Nonetheless, there is great variability in the stability of eukaryotic mRNAs: some mRNAs persist for only a few minutes, whereas others last for hours, days, or even months. These variations can produce large differences in the amount of protein that is synthesized.
Various factors, including the 5′ cap and the poly(A) tail (see Chapter 10), affect the stability of eukaryotic mRNA. Poly(A)-binding proteins (PABPs) normally bind to the poly(A) tail and contribute to its stability-
Much of RNA degradation in eukaryotes takes place in specialized complexes called P bodies. P bodies help control the expression of genes by regulating which RNA molecules are degraded and which are sequestered for later release. RNA degradation facilitated by small interfering RNAs (see next section) can also take place within P bodies.
Other parts of eukaryotic mRNA, including sequences in the 5′ untranslated region (5′ UTR), the coding region, and the 3′ UTR, also affect mRNA stability. Some short-
CONCEPTS
The stability of mRNA influences gene expression by affecting the amount of mRNA available to be translated. The stability of mRNA is affected by the 5′ cap, the poly(A) tail, the 5′ UTR, the coding region, and sequences in the 3′ UTR.
 CONCEPT CHECK 9
CONCEPT CHECK 9
How does the poly(A) tail affect mRNA stability?
The poly(A) tail stabilizes the 5′ cap, which must be removed before the mRNA molecule can be degraded from the 5′ end.