Molecular Mechanisms of Epigenetic Changes
Most evidence suggests that epigenetic effects are brought about by physical changes to chromatin structure. We will consider three types of molecular mechanisms that alter chromatin structure and underlie many epigenetic phenotypes: (1) changes in patterns of DNA methylation; (2) chemical modifications of histone proteins; and (3) RNA molecules that affect chromatin structure and gene expression.
DNA METHYLATION The best-
The fact that epigenetic changes are passed to other cells and (sometimes) to future generations means that the changes in chromatin structure associated with epigenetic phenotypes must be faithfully maintained when chromosomes replicate. How are epigenetic changes retained and replicated through the process of cell division?
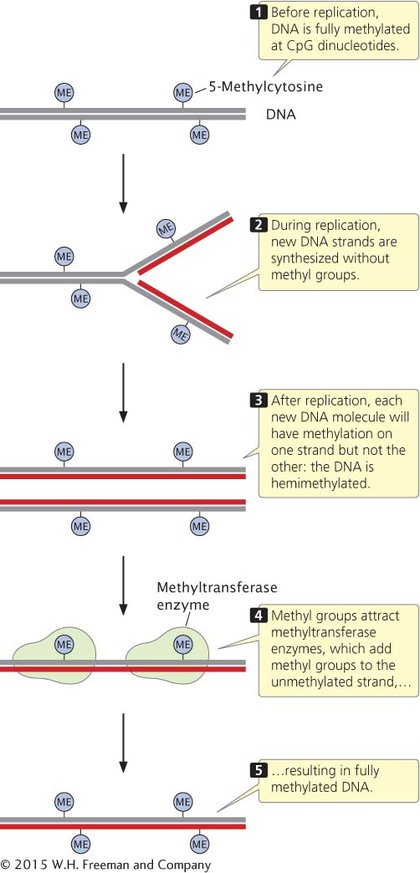
Methylation of CpG dinucleotides means that two methylated cytosine bases sit diagonally across from each other on opposite strands. Before replication, cytosine bases on both strands are methylated (Figure 12.23). Immediately after semiconservative replication, the cytosine base on the template strand has a methyl group, but the cytosine base on the newly replicated strand does not. Special methyltransferase enzymes recognize the hemimethylated state of CpG dinucleotides and add methyl groups to the unmethylated cytosine bases, resulting in two new DNA molecules that are fully methylated. In this way, the methylation pattern of DNA is maintained across cell division.
HISTONE MODIFICATIONS Epigenetic changes can also occur through modification of histone proteins. As we saw in Section 12.3, these modifications can alter chromatin structure and affect the transcription of genes. These types of modifications have been called epigenetic marks. For example, the addition of three methyl groups to lysine 4 in the H3 histone (H3K4me3) is often found near transcriptionally active genes. Methylation of lysine 36 in the H3 histone (H3K36me3) is also associated with increased transcription. On the other hand, the addition of three methyl groups to lysine 9 in H3 (H3K9me3) and to lysine 20 in histone 4 (H4K20me3) is associated with repression of transcription. Many additional histone modifications have been shown to be associated with the level of transcription.
Several models have been proposed to explain how histone modifications are faithfully transmitted to daughter cells. During the process of DNA replication, nucleosomes are disrupted and the original histone proteins are distributed randomly between the two new DNA molecules. Newly synthesized histones are then added to complete the formation of new nucleosomes (see Chapter 8). Most models assume that after replication, the epigenetic marks remain on the original histones; these marks then recruit enzymes that make similar changes to the new histones, maintaining the histone modifications across cell division.
EPIGENETIC EFFECTS OF RNA MOLECULES Evidence increasingly demonstrates that RNA molecules play an important role in bringing about epigenetic effects. The first discovered and still best-
CONCEPTS
DNA methylation, histone modifications, and RNA molecules bring about alterations of chromatin structure. Some of these modifications are passed to daughter cells during cell division and to future generations, producing epigenetic effects.