Spontaneous Replication Errors
Replication is amazingly accurate: less than one error in a billion nucleotides arises in the course of DNA synthesis (see Chapter 9). However, spontaneous replication errors do occasionally occur.
TAUTOMERIC SHIFTS The primary cause of spontaneous replication errors was formerly thought to be tautomeric shifts, in which the positions of protons in the DNA bases change. Each of the four bases exists in different chemical forms called tautomers. The two tautomeric forms of each base are in dynamic equilibrium, although one form is much more common than the other. The standard Watson-
Watson and Crick proposed that tautomeric shifts might produce mutations, and for many years their proposal was the accepted model for spontaneous replication errors. However, there has never been convincing evidence that the rare tautomers are the cause of spontaneous mutations. Furthermore, research now shows little evidence of tautomers in DNA.
MISPAIRING DUE TO OTHER STRUCTURES Mispairings often arise through wobble (see Chapter 11), in which normal, protonated, and other forms of the bases are able to pair because of flexibility in the DNA helical structure (Figure 13.10). These structures have been detected in DNA molecules and are now thought to be responsible for many of the mispairings in replication.
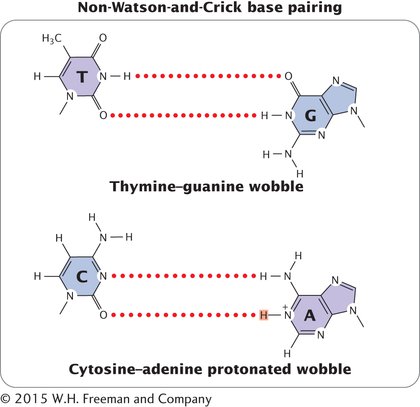
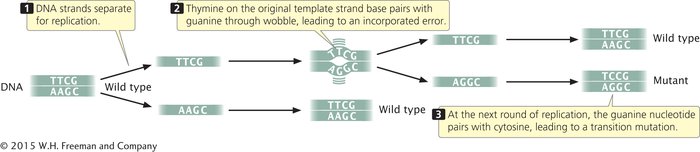
INCORPORATED ERRORS AND REPLICATED ERRORS When a mispaired base has been incorporated into a newly synthesized nucleotide chain, an incorporated error is said to have occurred. Suppose that, in replication, thymine (which normally pairs with adenine) mispairs with guanine through wobble (Figure 13.11). In the next round of replication, the two mismatched bases separate, and each serves as template for the synthesis of a new nucleotide strand. This time, thymine pairs with adenine, producing another copy of the original DNA sequence. On the other strand, however, the incorrectly incorporated guanine serves as the template and pairs with cytosine, producing a new DNA molecule that has an error: a C • G pair in place of the original T • A pair (a T • A → C • G base substitution). The original incorporated error leads to a replicated error, which creates a permanent mutation because all the base pairings are correct and there is no mechanism for repair systems to detect the error.
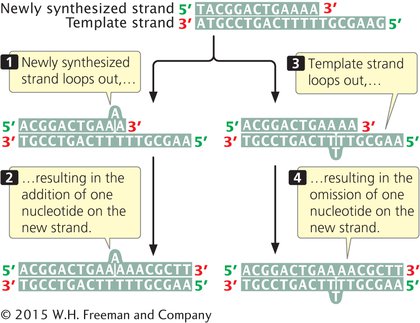
CAUSES OF DELETIONS AND INSERTIONS Small insertions and deletions can arise spontaneously in replication and crossing over. Strand slippage can occur when one nucleotide strand forms a small loop (Figure 13.12). If the looped-
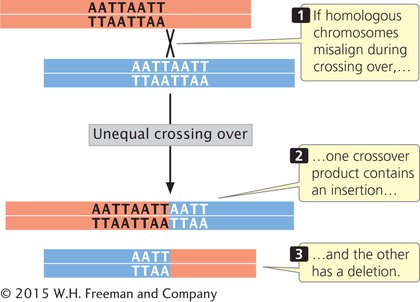
Another process that produces insertions and deletions is unequal crossing over. In normal crossing over, the homologous sequences of the two DNA molecules align, and crossing over produces no net change in the number of nucleotides in either molecule. Misaligned pairing can cause unequal crossing over, which results in one DNA molecule with an insertion and the other with a deletion (Figure 13.13).
CONCEPTS
Spontaneous replication errors arise from altered base structures and from wobble. Small insertions and deletions can occur through strand slippage in replication and through unequal crossing over.