DNA Sequencing
A powerful molecular method for analyzing DNA is a technique known as DNA sequencing, which determines the sequence of bases in DNA. Sequencing allows the genetic information in DNA to be read, providing an enormous amount of information about gene structure and function. In the mid-
The Sanger, or dideoxy, method of DNA sequencing is based on replication. The fragment to be sequenced is used as a template to make a series of new DNA molecules. In the process, replication is sometimes terminated when a specific base is encountered, producing DNA strands of different lengths, each of which ends in the same base.
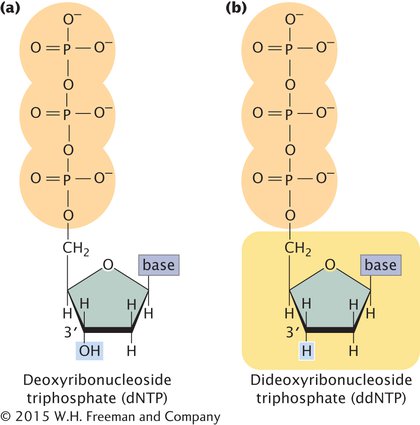
The method relies on the use of a special substrate for DNA synthesis. Normally, DNA is synthesized from deoxyribonucleoside triphosphates (dNTPs), which have an OH group on the 3′-carbon atom (Figure 14.13a). In the Sanger method, a special nucleotide, called a dideoxyribonucleoside triphosphate (ddNTP; Figure 14.13b), is also used as a substrate. The ddNTPs are identical to dNTPs, except that they lack a 3′-OH group. In the course of DNA synthesis, ddNTPs are incorporated into a growing DNA strand. However, after a ddNTP has been incorporated into the DNA strand, no more nucleotides can be added, because there is no 3′-OH group to form a phosphodiester bond with an incoming nucleotide. Thus, ddNTPs terminate DNA synthesis.
Although the sequencing of a single DNA molecule is technically possible, most sequencing procedures in use today require a considerable amount of DNA; any DNA fragment to be sequenced must first be amplified by PCR or by cloning in bacteria. Copies of the target DNA are then isolated and split into four samples (Figure 14.14). Each sample is placed in a different test tube to which the following ingredients are added:
Many copies of a primer that is complementary to one end of the target DNA strand
All four types of deoxyribonucleoside triphosphates, the normal precursors of DNA synthesis
A small amount of one of the four types of dideoxyribonucleoside triphosphates (ddATP, ddCTP, ddGTP, or ddTTP), which will terminate DNA synthesis as soon as it is incorporated into any growing chain (each of the four tubes receives a different ddNTP)
DNA polymerase
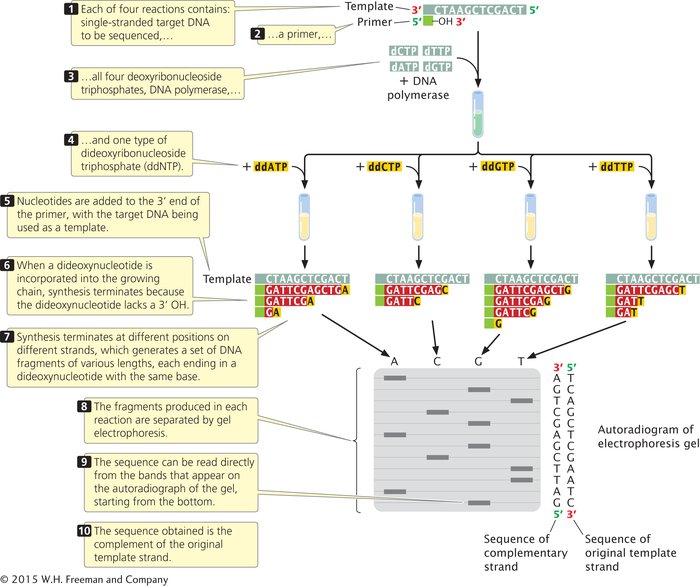
Either the primer or the dNTPs are radioactively or chemically labeled so that newly produced DNA can be detected.
Within each of the four tubes, the DNA polymerase synthesizes DNA. Let’s consider the reaction in one of the four tubes: the one that received ddATP. Within this tube, each of the single strands of target DNA serves as a template for DNA synthesis. The primer pairs with its complementary sequence at one end of each template strand, providing a 3′-OH group for the initiation of DNA synthesis. DNA polymerase elongates a new strand of DNA from this primer. Wherever DNA polymerase encounters a T on the template strand, it uses, at random, either a dATP or a ddATP to introduce an A into the newly synthesized strand. Because there is more dATP than ddATP in the reaction mixture, dATP is incorporated most often, allowing DNA synthesis to continue. Occasionally, however, ddATP is incorporated into the strand, and synthesis terminates. The incorporation of ddATP into the new strand takes place randomly at different positions in different copies, producing a set of DNA fragments of different lengths (12, 7, and 2 nucleotides long in the example illustrated in Figure 14.14), each ending in a nucleotide that contains adenine.
Equivalent reactions take place in the other three tubes, except that synthesis is terminated at nucleotides with a different base in each tube. After the completion of the polymerization reactions, all the DNA in the tubes is denatured, and the single-
The contents of the four tubes are separated side by side on an acrylamide gel (which allows finer separation than agarose) so that DNA strands differing in length by only a single nucleotide can be distinguished. After electrophoresis, the locations, and therefore the sizes, of the DNA strands in the gel are revealed by the radioactive or chemical labels.
Reading the DNA sequence is the simplest and shortest part of the procedure. In Figure 14.14, you can see that the band closest to the bottom of the gel is from the tube that contained the ddGTP reaction, which means that the first nucleotide added was guanine (G). The next band up is from the tube that contained ddATP; so the next nucleotide in the sequence is adenine (A), and so forth. In this way, the sequence is read from the bottom to the top of the gel, with the nucleotides near the bottom corresponding to the 5′ end of the newly synthesized DNA strand and those near the top corresponding to the 3′ end. Keep in mind that the sequence obtained is not that of the target DNA, but that of its complement. To see dideoxy sequencing in action, view Animation 19.3.  TRY PROBLEM 26
TRY PROBLEM 26
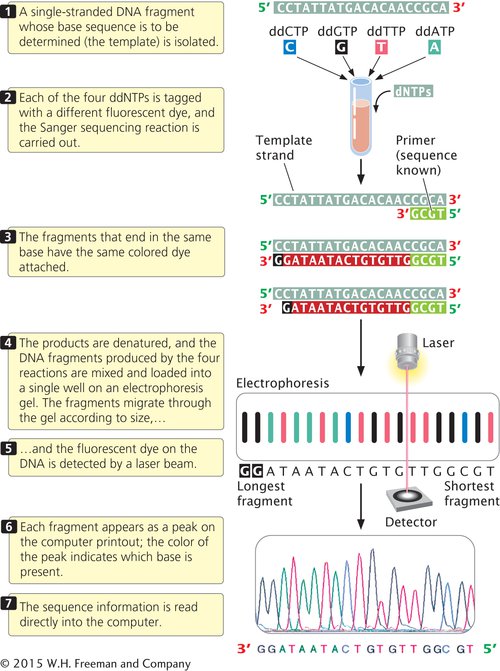
For many years, DNA sequencing was done largely by hand and was laborious and expensive. Today, sequencing is usually carried out by automated machines that use fluorescent dyes and laser scanners to sequence thousands of base pairs in a few hours (Figure 14.15). The dideoxy reaction is still used, but the ddNTPs used in the reaction are labeled with a fluorescent dye, and a different-
CONCEPTS
The dideoxy sequencing method uses ddNTPs, which terminate DNA synthesis at specific bases.
 CONCEPT CHECK 4
CONCEPT CHECK 4
In the dideoxy sequencing reaction, what terminates DNA synthesis at a particular base?
The absence of a base on the ddNTP halts the DNA polymerase.
The ddNTP causes a break in the sugar–phosphate backbone.
DNA polymerase will not incorporate a ddNTP into the growing DNA strand.
The absence of a 3′-OH group on the ddNTP prevents the addition of another nucleotide.
d