DNA Fingerprinting
The use of DNA sequences to identify individual persons is called DNA fingerprinting or DNA profiling. Because some parts of the genome are highly variable, each person’s DNA sequence is unique and, like a traditional fingerprint, provides a distinctive characteristic that allows identification.
Today, most DNA fingerprinting uses microsatellites, or short tandem repeats (STRs), which are very short DNA sequences repeated in tandem (see Section 8.6). These repeated sequences are found at many loci throughout the human genome. People vary in the number of copies of repeat sequences they possess at each of these loci. The STRs are typically detected with PCR, using primers flanking the repeats so that a DNA fragment containing the repeated sequences is amplified (Figure 14.18). The length of the amplified segment depends on the number of repeats; DNA from a person with more repeats will produce a longer amplified segment than will DNA from a person with fewer repeats.
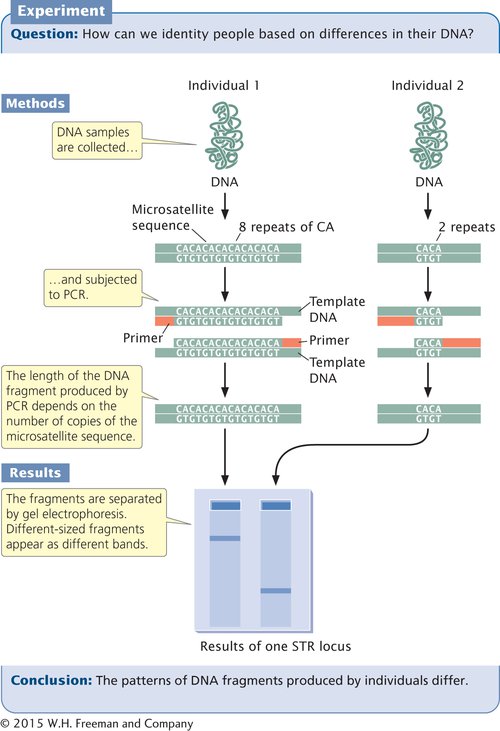
In DNA fingerprinting, the primers used in the PCR reaction are tagged with fluorescent labels so that the resulting DNA fragments can be detected with a laser. Primers for different STR loci are given different-
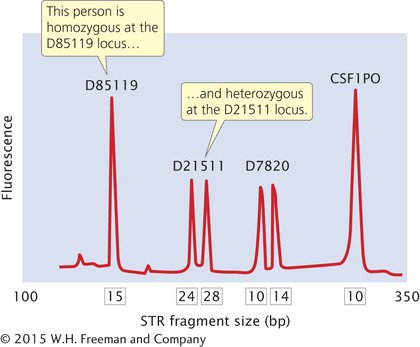
The Federal Bureau of Investigation has developed a system of 13 STR loci (Table 14.3) that are commonly used for identifying people and solving crimes. These loci make up the Combined DNA Index System (CODIS). Each STR locus in CODIS has a large number of alleles and is located on a different human chromosome, so variation at each locus assorts independently. When all 13 CODIS loci are used together, the probability of two randomly selected people having the same DNA profile is less than 1 in 10 billion.
| Locus name | Chromosome | Number of repeats | Number of alleles* |
|---|---|---|---|
| CSF1PO | 5 | 5–17 | 10 |
| FGA | 4 | 12–51 | 23 |
| TH01 | 11 | 3–14 | 8 |
| TPOX | 2 | 4–16 | 8 |
| VWA | 12 | 10–25 | 10 |
| D3S1358 | 3 | 6–26 | 10 |
| D5S818 | 5 | 4–29 | 9 |
| D7S820 | 7 | 5–16 | 11 |
| D8S1179 | 8 | 6–20 | 11 |
| D13S317 | 13 | 5–17 | 8 |
| D16S539 | 16 | 4–17 | 7 |
| D18S51 | 18 | 5–40 | 19 |
| D21S11 | 21 | 12–43 | 22 |
|
Source: Data from J. M. Butler and C. R. Hill, Forensic Science Review 24:15–26, 2012. *U.S. population. |
|||
In a typical application, DNA fingerprinting might be used to confirm that a suspect was present at the scene of a crime. A sample of DNA from blood, semen, hair, or other body tissue is collected from the crime scene. If the sample is very small, PCR can be used to amplify it so that enough DNA is available for testing. Additional DNA samples are collected from one or more suspects. The pattern of DNA fragments produced by DNA fingerprinting of the sample is then compared with the patterns produced by DNA fingerprinting of the suspects. A match between the sample and a suspect can provide evidence that the suspect was present at the scene of the crime (Figure 14.20).
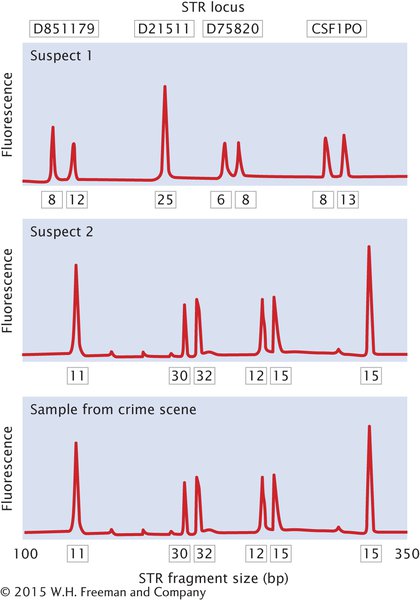
Since its introduction in the 1980s, DNA fingerprinting has helped convict a number of suspects in murder and rape cases. Suspects in other cases have been proved innocent when their DNA failed to match that from the crime scenes. Initially, calculations of the odds of a match (the probability that two people could have the same pattern) were controversial, and there were concerns about quality control (such as the accidental contamination of samples and the reproducibility of results) in laboratories where DNA analysis was done. Today, DNA fingerprinting has become widely accepted as an important tool in forensic investigations. In addition to its application in solving crimes, DNA fingerprinting is used to assess paternity, study genetic relationships among individual organisms in natural populations, identify specific strains of pathogenic bacteria, and identify human remains. For example, DNA fingerprinting was used to determine that several samples of anthrax mailed to different people in 2001 were all from the same source.
CONCEPTS
DNA fingerprinting detects genetic differences among people by analyzing highly variable regions of chromosomes.
 CONCEPT CHECK 5
CONCEPT CHECK 5
How are microsatellites detected?
Microsatellites are detected by using PCR with primers that flank the region containing the repeats.