WORKED PROBLEM
WORKED PROBLEM
Problem 1
A molecule of double-
BamHI (recognition sequence is GGATCC)
HindIII (recognition sequence is AAGCTT)
HpaII (recognition sequence is CCGG)
Solution Strategy
What information is required in your answer to the problem?
The number of restriction sites likely to be present in the DNA molecule for each of the specified restriction enzymes.
What information is provided to solve the problem?
The size of the DNA molecule.
The G + C base composition of the DNA molecule.
The recognition sequences for each restriction enzyme.
For help with this problem, review:
Cutting and Joining DNA Fragments in Section 14.1.
Hint: If you know the percentage of any base in the DNA, you can determine the percentages of all the other bases, because G = C and A = T.
Remember: The multiplication rule states that the probability of two or more independent events is calculated by multiplying their independent probabilities.
Solution Steps
The percentages of G and C are equal in double-
The probability of finding the sequence GGATCC = 0.31 × 0.31 × 0.19 × 0.19 × 0.31 × 0.31 = 0.0003333. To determine the average number of recognition sequences in a 5 million-
base- pair piece of DNA, we multiply 5,000,000 bp × 0.00033 = 1666.5 recognition sequences. The number of AAGCTT recognition sequences is 0.19 × 0.19 × 0.31 × 0.31 × 0.19 × 0.19 × 5,000,000 = 626 recognition sequences.
The number of CCGG recognition sequences is 0.31 × 0.31 × 0.31 × 0.31 × 5,000,000 = 46,176 recognition sequences.
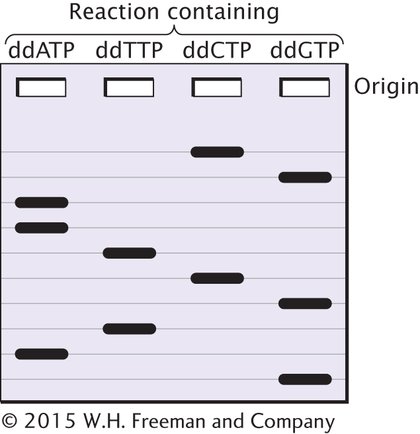
Problem 2
You are given the following DNA fragment to sequence: 5′—GCTTAGCATC—
Solution Strategy
What information is required in your answer to the problem?
The positions of the bands on the sequencing gel.
What information is provided to solve the problem?
The base sequence of the DNA fragment to be sequenced.
For help with this problem, review:
Remember: In dideoxy sequencing, a new DNA strand is synthesized, and that strand is what is sequenced. Thus, the bands that appear on the gel represent the complement of the original sequence.
DNA Sequencing in Section 14.3.
Solution Steps
The first task is to write out the sequence of the newly synthesized fragment, which will be complementary and antiparallel to the original fragment. The original sequence is 5′—GCTTAGCATC—
Hint: Small fragments, those nearer the 5′ end of the newly synthesized strand, will migrate faster and will appear near the bottom of the gel.
3′—CGAATCGTAG—
Written 5′→3′, the sequence is 5′—GATGCTAAGC—