Single-Nucleotide Polymorphisms
Since the completion of the sequencing of the human genome, sequencers have focused much of their effort on mapping differences among people in their genomic sequences.
Imagine that you are riding in an elevator with a random stranger. How much of your genome do you have in common with this person? Studies of variation in the human genome indicate that you and the stranger will be identical at about 99.9% of your DNA sequences. This difference is very small in relative terms, but because the human genome is so large (3.2 billion base pairs), you and the stranger will actually be different at more than 3 million base pairs of your genomic DNA. These differences are what make each of us unique, and they greatly affect our physical features, our health, and possibly even our intelligence and personality.
A site in the genome at which individual members of a species differ in a single base pair is called a single-nucleotide polymorphism (SNP, pronounced “snip”). Arising through mutation, SNPs are inherited as allelic variants (just as alleles that produce phenotypic differences, such as blood types, are, although SNPs do not usually produce phenotypic differences). Single-nucleotide polymorphisms are numerous and are present throughout genomes. In a comparison of the same chromosome from two different people, a SNP can be found approximately every 1000 bp.
Most SNPs present within a population arose once from a single mutation that occurred on a particular chromosome and subsequently spread throughout the population. Thus, each SNP is initially associated with other SNPs (as well as with other types of genetic variants or alleles) that were present on the particular chromosome on which the mutation arose. The specific set of SNPs and other genetic variants observed on a single chromosome or part of a chromosome is called a haplotype (Figure 15.6). Single-nucleotide polymorphisms within a haplotype are physically linked and therefore tend to be inherited together. New haplotypes can arise through mutation or crossing over, which breaks up the particular set of SNPs in a haplotype.
Page 410
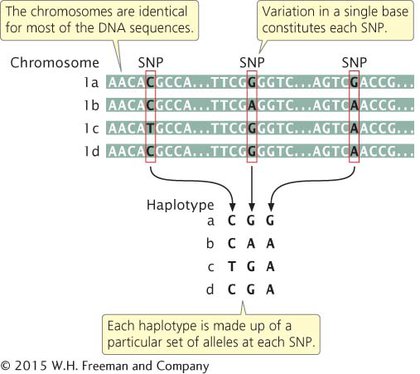
15.6 A haplotype is a specific set of single-nucleotide polymorphisms (SNPs) and other genetic variants observed on a single chromosome or part of a chromosome. Chromosomes 1a, 1b, 1c, and 1d represent different copies of a chromosome that might be found in a population.
Because of their variability and widespread occurrence throughout the genome, SNPs are valuable as markers in linkage studies. When a SNP is physically close to a disease-causing locus, it will tend to be inherited along with the disease-causing allele. Therefore, people with the disease will tend to have different SNPs than healthy people. A comparison of SNP haplotypes in people with the disease and in healthy people can reveal the presence of genes that affect the disease; because the disease gene and the SNP are closely linked, the location of the disease-causing gene can be determined from the locations of associated SNPs. This approach is the same as that used in gene mapping with RFLPs (see Chapter 14), but there are many more SNPs than RFLPs, providing a dense set of variable markers covering the entire genome that can be used more effectively in mapping.
GENOME-WIDE ASSOCIATION STUDIES Many common diseases are caused by complex interactions among multiple genes; the availability of SNPs has greatly facilitated the search for these genes. Genome-wide association studies use numerous SNPs scattered across the genome to find genes of interest. In one of the most successful applications of SNPs for finding disease associations, researchers genotyped 17,000 people in the United Kingdom for 500,000 SNPs in 2007. They detected strong associations between 24 genes or chromosome segments and the incidence of seven common diseases, including coronary artery disease, Crohn disease, rheumatoid arthritis, bipolar disorder, hypertension, and two types of diabetes. The importance of this study is its demonstration that genome-wide association studies using SNPs can successfully locate genes that contribute to complex diseases caused by multiple genetic and environmental factors.
Within the past few years, SNPs have been used in genome-wide association studies to successfully locate genes that influence many additional traits, such as the age of puberty and menopause in women, variation in facial features, skin pigmentation, eye color, body weight, bone density, glaucoma, and even susceptibility to infectious diseases such as meningococcal disease and tuberculosis. Unfortunately, the genes identified often explain only a small proportion of the genetic influence on the trait. For example, one huge genome-wide association study combined data from over 100,000 human subjects in an attempt to locate genes encoding blood lipids involved in cardiovascular disease. Although the study identified 95 different loci associated with lipid traits, those genes corresponded to only 25%–30% of the total genetic variation in these traits. DNA sequences that encode the majority of the missing genetic variation in such traits—sometimes called the “dark matter of the genome”—have thus far remained largely undetected. The low percentage of variation explained by most current genome-wide association studies means that the genes identified are not, by themselves, useful predictors of the risk of inheriting the disease or trait. Nevertheless, the identification of specific genes that influence a disease or trait can lead to a better understanding of the biological processes that produce the phenotype.
