Testing for Hardy–Weinberg Proportions
To determine if a population’s genotypes are in Hardy–Weinberg equilibrium, the genotypic proportions expected under the Hardy–Weinberg law must be compared with the observed genotypic frequencies. To do so, we first calculate the allelic frequencies, then find the expected genotypic frequencies by using the square of the allelic frequencies, and finally, compare the observed and expected genotypic frequencies by using a chi-
WORKED PROBLEM
Jeffrey Mitton and his colleagues found three genotypes (R2R2, R2R3, and R3R3) at a locus encoding the enzyme peroxidase in ponderosa pine trees growing at Glacier Lake, Colorado. The observed numbers of these genotypes were
| Genotypes | Number observed |
|---|---|
| R2R2 | 135 |
| R2R3 | 44 |
| R3R3 | 11 |
Are the ponderosa pine trees at Glacier Lake in Hardy–Weinberg equilibrium at the peroxidase locus?
Solution Strategy
What information is required in your answer to the problem?
The results of a chi-
What information is provided to solve the problem?
The numbers of the different genotypes in a sample of the population.
Solution Steps
If the frequency of the R2 allele equals p and the frequency of the R3 allele equals q, the frequency of the R2 allele is
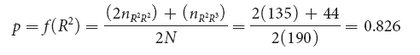
The frequency of the R3 allele is obtained by subtraction:
q = f(R3) = 1 – p = 0.174
The frequencies of the genotypes expected under Hardy–Weinberg equilibrium are then calculated by using p2, 2pq, and q2:
R2R2 = p2 = (0.826)2 = 0.683
R2R3 = 2pq = 2(0.826)(0.174) = 0.287
R3R3 = q2 = (0.174)2 = 0.03
Multiplying each of these expected genotypic frequencies by the total number of observed genotypes in the sample (190), we obtain the numbers expected for each genotype:
R2R2 = 0.683 × 190 = 129.8
R2R3 = 0.287 × 190 = 54.5
R3R3 = 0.03 × 190 = 5.7
By comparing these expected numbers with the observed numbers of each genotype, we see that there are more R2R2 and R3R3 homozygotes and fewer R2R3 heterozygotes in the population than we expect at equilibrium.
A chi-

The calculated chi-
So far, the chi-
After we have calculated both the chi-
 For additional practice, determine whether the genotypic frequencies in Problem 26 at the end of the chapter are in Hardy–Weinberg equilibrium.
For additional practice, determine whether the genotypic frequencies in Problem 26 at the end of the chapter are in Hardy–Weinberg equilibrium.
CONCEPTS
The observed number of genotypes in a population can be compared with the expected Hardy–Weinberg proportions by using a chi-